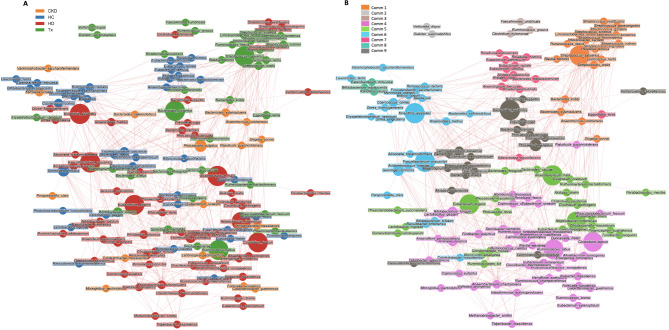
Fig. 6.
Network analysis shows clusters of microbial species and positive or negative correlations of their abundances. (A) Nodes (representing species) were colored according to the patient group in which species had the highest mean abundance. (B) Nodes were colored according to the detected communities using the Louvain algorithm. Node size is directly proportional to the centrality of the species within the overall network. Edge thickness is inversely proportional to the Pearson p-value (10% Benjamini–Hochberg two-stages FDR), and it is colored according to the positive (red) or negative (blue) Pearson’s coefficient. CKD: non-dialysis Chronic Kidney Disease, HD: Hemodialysis, Tx: Kidney transplant), HC: Healthy Controls, Comm: community.