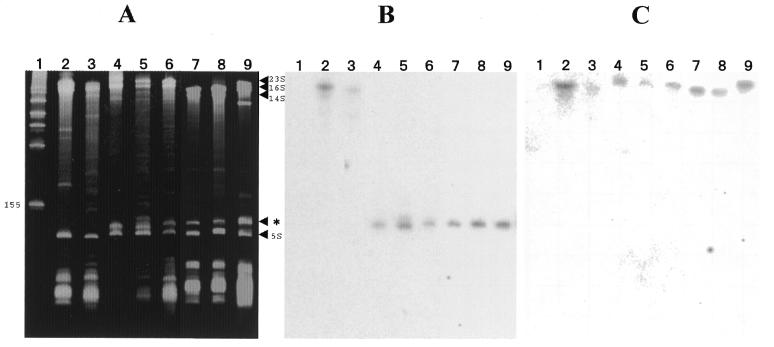
Figure 1.
Acrylamide gel electrophoresis and northern hybridization analysis of RNA from E.coli and α-proteobacteria. Lane 1, marker RNA; lane 2, E.coli total RNA; lane 3, R.rubrum total RNA; lane 4, R.palustris purified LSU ribosomes; lane 5, USDA 4362 total RNA; lane 6, USDA 4377 total RNA; lane 7, R.sphaeroides total RNA; lane 8, R.capsulatus total RNA; lane 9, M.extorquens total RNA. (A) 6.5% acrylamide denaturing gel. Positions of 23S rRNA, 16S rRNA, 14S rRNA and 5S rRNA are indicated on the right margin as well as the 23S rRNA 5′ domain (*). Position of the lowest molecular weight marker band is also indicated on the left margin (155 nt). (B) Northern hybridization of a 5′-32P-labeled oligonucleotide complementary to positions 44–61 in R.palustris 23S rRNA to RNA transferred to a nitrocellulose filter from the gel in (A). (C) Northern hybridization of a 5′-32P-labeled oligonucleotide complementary to positions 503–523 in R.palustris 23S rRNA to RNA transferred to a nitrocellulose filter from a gel identical to that in (A). 23S rRNA sequences chosen for synthesis of complementary oligonucleotides in experiments of (B) and (C) are completely conserved in all of the bacterial 23S rRNAs studied here.