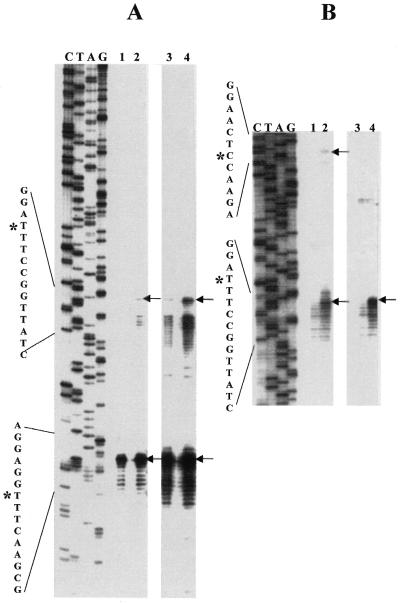
Figure 4.
(A) Primer extension identifies the right side of the Bradyrhizobial IVS and also the extent of the IPE. Primer extension analysis of total RNA from phototrophic Bradyrhizobium USDA 4362 and USDA 4377 using a 5′-32P-labeled primer at 23S rRNA positions 305–325 complementary to the R.palustris 23S rRNA sequence. Lefthand lanes (C, T, A, G) are dideoxy sequence marker generated with the same primer and 23S rDNA-containing plasmid DNA from USDA 4377. Since the sequences of USDA 4362 and USDA 4377 are identical in this region, only the USDA 4377 dideoxy sequence marker is presented. Lanes 1 and 2 from the same gel are intentionally overexposed (lanes 3 and 4) to show the presence of low abundance rRNA 5′-ends derived from partially processed 23S rRNA intermediates. DNA sequence around the processing sites is presented on the left margin, with position of the cleavage which generates the first detectable intermediate (*) on the upper sequence and the predominant cleavage which generates completely processed 23S rRNA (*) on the lower sequence. (B) Primer extension was performed on a mixture of rRNA precursors partially processed in vitro with E.coli RNase III. From Bradyrhizobium USDA 4377 (lanes 1 and 2) and Bradyrhizobium USDA 4362 (lanes 3 and 4). In lanes 1 and 3, the RNA processing reactions were terminated immediately by phenol extraction while in lanes 2 and 4, processing reactions were allowed to proceed for 30 min. The first detectable in vivo cleavage [upper arrows in (A)] corresponds precisely to the site cleaved in vitro by RNase III on the right side of stem–loop structure #1 in the Bradyrhizobium USDA 4362 and USDA 4377 rRNA precursors [lower arrows in (B)]. In the case of the Bradyrhizobium USDA 4362 RNase III processing reaction, primer extension products corresponding to the cleavage at the left side of stem–loop structure #1 migrate more slowly and have not entered this portion of the gel.