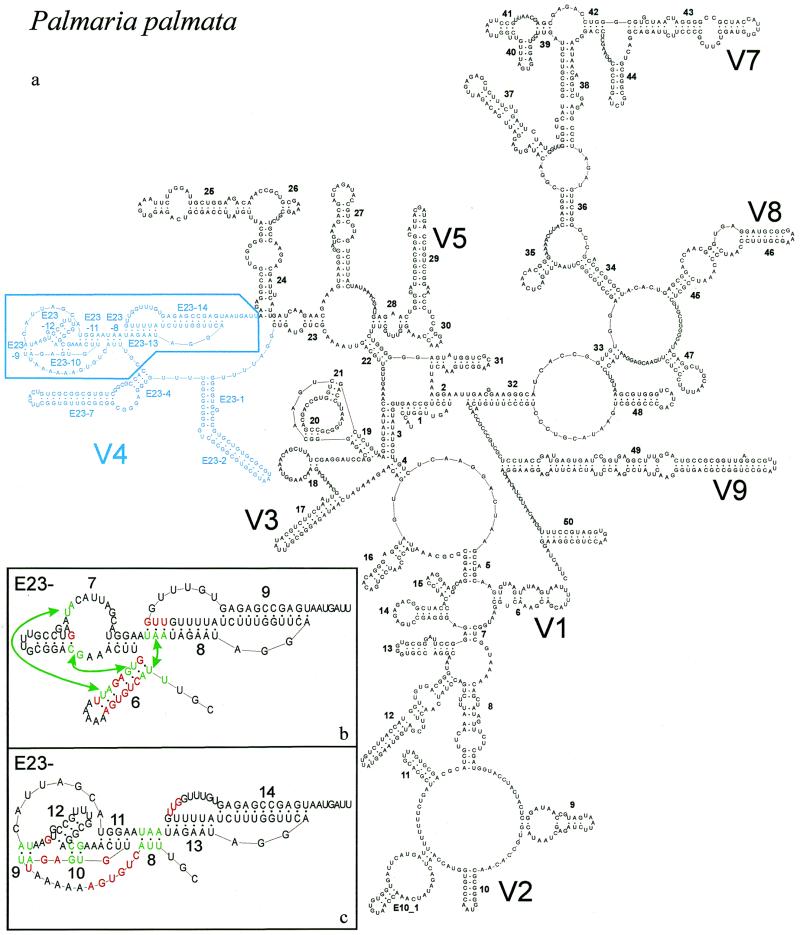
Figure 1.
New secondary structure model illustrated with P.palmata SSU rRNA. (a) Complete model with indication of variable areas V1–V9. V6 is variable only in prokaryotic SSU rRNA where helix 37 is branched. Area V4 is in blue; base pair compensation and base covariation was examined systematically for the boxed part, details of which are shown in the inserts (b) and (c). (b) Old model and helix numbering for part of area V4. Abolished base pairs are in red and new ones, connected by arrows, are in green. (c) New model and helix numbering for part of area V4. Color conventions as in (b). The numbering system was changed to fit the new helix succession and to allow numbering of extra helices present in certain taxa (see Table 3). The correspondence between old and new helix number is as follows: helix E23-6 of the old model is dismantled; E23-7 (old) is transformed into helices E23-11 and 12 (new); E23-8 (old) is partly conserved as E23-13 (new); E23-9 (old) becomes E23-14 (new). Helices E23-8,9 and 10 of the new model do not exist in the old model.