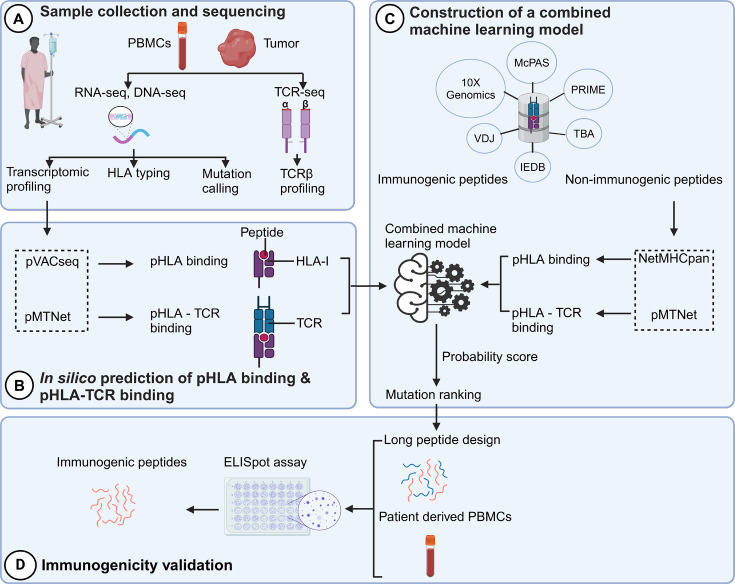
Figure 1. A novel workflow based on machine learning that integrates T cell receptor β (TCRβ) sequencing data for the identification and ranking of colorectal cancer (CRC) neoantigens.
(A) Tumor biopsies and peripheral blood from CRC patients were subjected to targeted DNA-seq, RNA-seq, and T cell receptor (TCR)-seq. (B) The prediction of peptide-human leukocyte antigen (HLA) binding and peptide-HLA-TCR binding by indicated tools using the DNA-seq, RNA-seq, and TCR-seq data was performed. (C) Machine learning models were subsequently constructed based on the analysis of the peptide-HLA binding and peptide-HLA-TCR binding features to distinguish immunogenic antigens from non-immunogenic peptides. (D) The immunogenicity of predicted neoantigen candidates prioritized by the model was validated by enzyme-linked immunospot (ELISpot) to evaluate the effectiveness of this approach.