Figure 1.
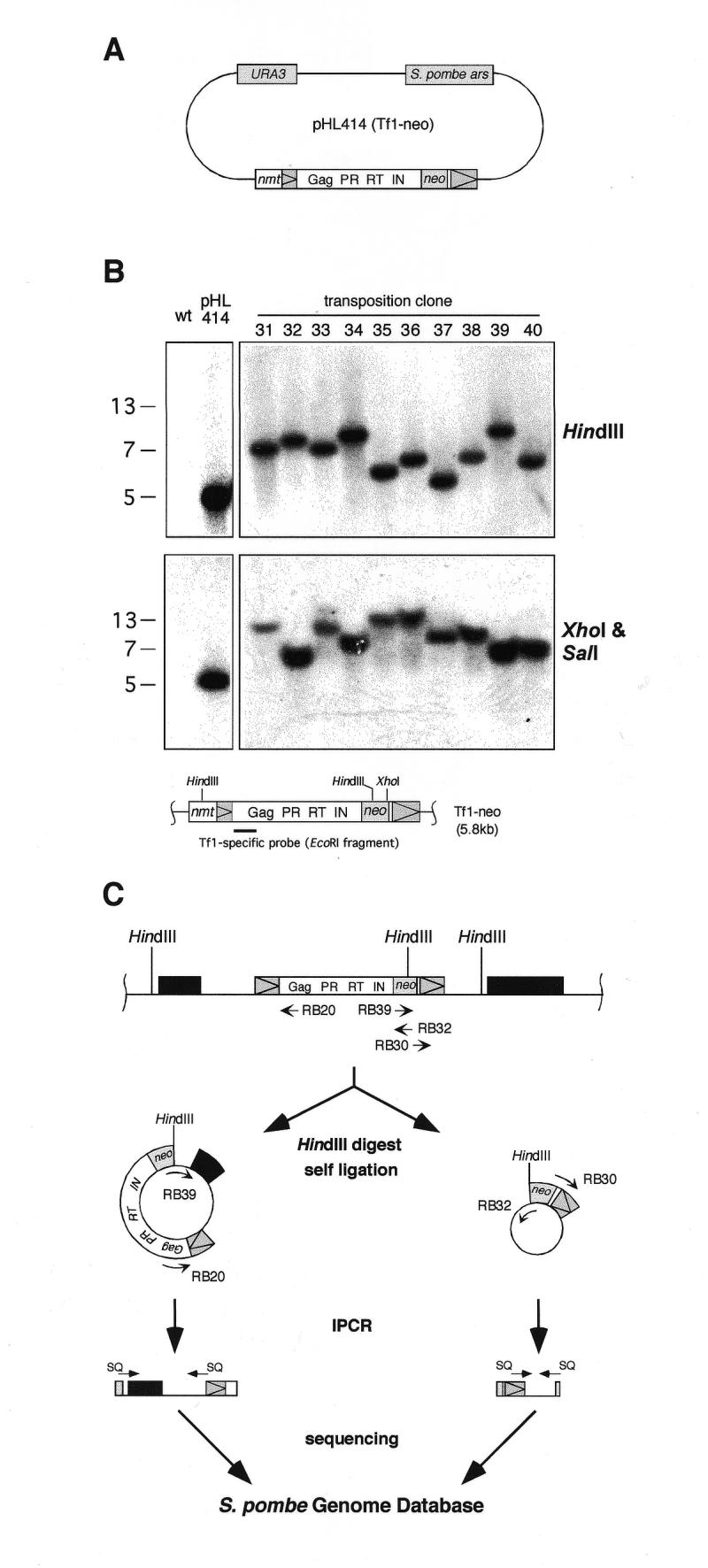
Determination of genomic Tf1-neo insertions. (A) The transposition assay plasmid pHL414 contained the URA3 gene of S.cerevisiae and the Tf1-neo element fused to an inducible nmt promoter. The neo gene inserted in Tf1 confers G418 resistance. The large white box represents the single full-length ORF of Tf1. The positions of the protein domains Gag, protease (PR), reverse transcriptase (RT) and integrase (IN) are indicated. Triangles within grey boxes represent LTRs. The plasmid contains an autonomous replication sequence of S.pombe (ars). (B) Southern blot analysis of Tf1 transposition clones. DNA from G418-resistant clones was cut with restriction enzymes as indicated, blotted, and hybridised to a Tf1-specific probe shown in the schematic presentation of the nmt–Tf1-neo fusion construct. As controls, DNA from an isogenic strain without Tf1 insertion (wt) and from the same strain carrying the pHL414 plasmid (pHL414) in the presence of thiamine were used. Of 40 analysed transposition clones, 10 representative clones are shown here. Molecular weights are shown to the left of the gel. (C) The genomic locations of induced Tf1-neo insertion events were determined using IPCR and the S.pombe genome database. IPCR primers (see Materials and Methods; RB30, RB32: primer set 1; RB20, RB39: primer set 2) and sequencing primers (SQ) are shown as arrows. HindIII restriction sites are indicated, and genes are represented as black boxes.
