Abstract
The Serratia marcescens hemophore-specific outer membrane receptor HasR is a member of the TonB-dependent family of autoregulated receptors. It can transport either heme itself or heme bound to the hemophore HasA. On the basis of sequence and functional similarities with other TonB-dependent outer membrane receptors whose three-dimensional structures have been determined, a HasR structure model was proposed. The mature HasR protein comprises a 99-residue amino-terminal extension necessary for hasR transcription, followed by a plug domain of 139 amino acids and a β-barrel domain inserted in the outer membrane, the lumen of which is closed by the plug. This model was used to generate hasR deletions encoding HasR proteins with the native signal peptides but lacking either the N-terminal regulatory extension or encoding the plug or the β-barrel alone. The protein lacking the N-terminal extension, HasR Δ11-91, was incorporated in the outer membrane and was fully functional for active uptake of free and hemophore-bound heme. The HasR β-barrel, Δ11-192, was also incorporated in the outer membrane and bound the hemophore but expressed no active heme transport properties. The HasR plug remained in the periplasm. Coexpression of the plug and the β-barrel allowed partial plug insertion in the outer membrane, demonstrating that these two HasR domains interact in vivo. The β-barrel and the plug also interact in vitro. Nevertheless, the two domains did not complement each other to reconstitute an active TonB-dependent receptor for free or hemophore-bound heme uptake. Production of the β-barrel alone selectively increased passive diffusion of heme but not of other exogenous compounds. A mutation at histidine 603, which is required for heme uptake through the wild-type receptor, abolished heme diffusion, showing that HasR Δ11-192 forms a specific heme channel.
Gram-negative bacteria are surrounded by an outer membrane outside the peptidoglycan layer which delimits the periplasmic space, confining many extracytoplasmic enzymes to that space. The outer membrane also serves as a selective permeation barrier: it restricts permeability to lipophilic compounds and contains channels for nutrients and ions required for growth. These channels are made up of outer membrane proteins. They can be schematically grouped into three classes: passive nonspecific diffusion pores, specific channels, and active transporters. Diffusion channels allow entry of hydrophilic solutes smaller than 600 Da. Specific channels also allow some flow of small hydrophilic compounds and in addition bind their cognate substrates and allow their diffusion at micromolar concentrations. The three-dimensional structures of several outer membrane porins and specific channels have been determined. They form trimeric β-barrel structures, each subunit having a constricted central lumen of 12 Å diameter (see reference 27 for review).
Larger compounds like siderophores and heme are actively transported through specific gated receptors. Despite the large diversity of their cognate ligands, these siderophore and heme receptors work similarly. Transport requires energy derived from the proton motive force, which is provided by a protein complex comprising the inner membrane proteins ExbB, ExbD, and TonB (the TonB complex). In many species, there is only one such complex, and it is shared by the various iron receptors produced by the species. The periplasmic C-terminal part of TonB interacts with a short conserved region named the TonB box located close to the N terminus of the receptors. The three-dimensional structures of six such receptors in gram-negative bacteria have been determined (7, 10, 11, 12, 16, 18). They are monomeric proteins with a β-barrel domain inserted in the outer membrane whose lumen is entirely closed by the N-terminal plug domain, which is inserted from the periplasmic side. Several substrates bind to residues both in the external loops of the β-barrel and in the plug. A current model is that the plug binds the ligand and physically pulls it inside during its unwinding in the periplasm (17). It has not been established whether the plug is ejected from the β-barrel during substrate transport, transiently leaving an open wide channel (up to 50 Å) in the outer membrane.
The outer membrane receptor genes belong to operons regulated by the iron-loaded Fur repressor, and hence are tightly regulated by iron availability. Several of these receptors are also positively auto-regulated by the binding of their specific ligand via extra cytoplasmic function sigma factors associated with membrane-bound anti-sigma factors. Ligand binding to its cognate receptor triggers the activation of the sigma factor and transcription of the receptor gene. Autoregulated receptors have an N-terminal extension 50 to 100 amino acids long, named the regulatory extension, between the signal peptide and the TonB box. The signaling cascade also requires the TonB complex (36).
The structures of other iron/heme outer membrane receptors have not yet been determined but have been predicted from sequence and function analogies to fold similarly (37).
Heme receptors have been subdivided into three categories. One group recognizes heme and the second recognizes host hemoproteins. The third group interacts with hemophores, proteins secreted by several gram-negative bacteria that capture free heme or extract heme from heme carrier proteins, owing to their higher affinity for heme, thereby making it available to hemophore-specific outer membrane receptors (37). The specificity for one type of heme-containing molecule is not absolute, and several hemoprotein receptors also bind heme. This is the case for the Serratia marcescens hemophore receptor HasR, which binds the hemophore HasA with high affinity (in the nanomolar range) and heme with lower affinity (in the micromolar range) and promotes uptake of both free and hemophore-bound heme.
Hemophore binding provides a better heme source as it allows growth at lower heme concentrations (19). Both heme and hemophore are recognized by the receptor, but the primary interaction between receptor and hemophore does not involve the heme moiety and is mediated by two peptide segments in the hemophore which form independent binding regions (21). Hemophore binding to the receptor is independent of the TonB complex, but heme transport (free or bound to hemophore) is dependent on the TonB complex. Empty hemophore release after heme extraction is also TonB-ExbB-ExbD dependent and is the step that involves the highest energy consumption (22). HasR belongs to the autoregulated receptor family and has an N-terminal 99-amino-acid extension necessary for autoregulation. Binding of the heme-loaded hemophore to HasR triggers a signaling cascade which activates a specific extracytoplasmic function sigma factor which leads to hasR transcription (3). HasR shares significant similarity with other heme and hemoprotein receptors, in particular the conserved FRAP/NPL sequences in the β-barrel and two conserved histidines, one in the plug at 189 and one in the barrel at 603 (4).
A model of the HasR structure has been proposed and we used it to suggest hasR deletions encoding HasR mutant proteins in which the HasR cleavable signal sequence is linked either to the plug or to the β-barrel domain or to a polypeptide lacking the N-terminal regulatory extension. These polypeptides were produced separately or together and their functions were analyzed in vivo and in vitro.
We report that in the absence of the plug, the β-barrel alone is inserted into the outer membrane and can bind the hemophore. However, it is not capable of TonB-dependent active transport as previously shown for FhuA and FepA (5, 36). It selectively increases entry of heme but not of other exogenous compounds.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this work are listed in Table 1.
TABLE 1.
Bacterial strains, cloning vectors, and recombinant plasmids used in this study
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| E. coli | ||
| POP3 | araD139 ΔlacU169 rpsL relA thi | Laboratory collection |
| TG1 | supE Δ(hsdM-mcrB)5 thi Δ(lac proAB) F′ (traΔ36 proA+B+lacIqZΔM15) | Laboratory collection |
| C600 | F−thr leu fhuA lacY thi supE | Laboratory collection |
| C600 exbB::Tn10 | F−thr leu fhuA lacY thi supE exbB::Tn10 | Laboratory collection |
| C600 ΔhemA::km | ΔhemA::km | Laboratory collection (19) |
| C600 ΔhemA::km exbB::Tn10 | ΔhemA::km exbB::Tn10 | Laboratory collection (22) |
| Plasmids | ||
| pACYC184 | Cmr | Laboratory collection |
| pNC1 | pSU19 tonB-exbB-exbD, Cmr | 8 |
| pHSG576 fhuAΔ5-160 | pSC101 fhuAΔ5-160, Cmr | 5 |
| pBAD24 | pBR322 araC, arabinose-inducible promoter, Ampr | Laboratory collection |
| pBAD33 | pACYC184 araC, arabinose-inducible promoter, Cmr | Laboratory collection |
| pSYC34PAM | pAM238 lactose-inducible hasADE, Spcr | 23 |
| pSYC134PAM | pAM238 lactose-inducible hasA, Spcr | 23 |
| pQE32 | pUC18, oligohistidine tag, Ampr | Qiagen |
| pBAD24-hasR | pBR322 araC, arabinose-inducible hasR, Ampr | 21 |
Media.
Hemin was obtained from Sigma Chemical Co. and dissolved immediately before use in a minimal volume of 0.1 N NaOH, centrifuged, and diluted with the appropriate buffer to the desired concentration. Tetradecyl-N,N-dimethyl-3-ammonio-1-propanesulfonate (ZW 3-14) was obtained from Calbiochem. To chelate iron, 0.2 mM of 2,2′-dipyridyl was added to LB medium. Δ-Aminolevulinic acid was added to LB medium (LBΔ) to a final concentration of 20 μg ml−1 for heme auxotroph growth. The following antibiotics were used: ampicillin, 100 μg/ml; spectinomycin, 50 μg/ml; chloramphenicol, 50 μg/ml; kanamycin, 50 μg/ml; and tetracycline, 10 μg/ml. All cultures were grown with aeration at 30°C or 37°C and optical density was measured at 600 nm (OD600).
Extraction and manipulation of plasmids.
Standard methods were used for isolation of plasmids, cloning, restriction map analysis, and transformation.
Production and purification of the HasA protein.
Apo-HasA wild-type was obtained from culture supernatants of strain POP3(pSYC34PAM) grown at 30°C in M9 medium with glycine and spectinomycin. The supernatant was concentrated by 65% ammonium sulfate precipitation and then extensively dialyzed against TN (50 mM Tris-HCl, pH 7.5, 100 mM NaCl). The heme content of the resulting protein samples was determined from the absorbance at the Soret band wavelength. Less than 0.5% (mol/mol) of the purified proteins were loaded with heme. The purity of the protein preparation as estimated from sodium dodecyl sulfate (SDS) gels was more than 99%.
Preparation of membrane, cytoplasmic, and periplasmic fractions and protein analysis.
Strain C600 carrying the various plasmids was grown in 20 ml of LB containing 0.02% arabinose at 37°C or in LB containing 0.02% arabinose and 0.4% maltose (to induce the maltose binding protein [MBP] which is used as a periplasmic marker). Cells were harvested by centrifugation for 10 min at 5,000 × g at 4°C when they reached an optical density of 1 at 600 nm and were washed once in 50 mM Tris-HCl, pH 7.5. Each pellet was resuspended in 500 μl of 50 mM Tris-HCl, pH 7.5, and cells were broken either by sonication or by one passage through a French press. After centrifugation for 10 min at 5,000 × g at 4°C to remove unbroken cells and aggregates, the preparations were incubated for 15 min at room temperature with 10 μg/ml RNase and DNase.
The crude membrane pellets were collected by centrifugation for one hour at 15,000 × g at 4°C in a microcentrifuge. The supernatant contained the total soluble fractions, and the pellet contained all membranes (19). A protocol derived from Schalk et al. was used to solubilize the inner and outer membranes sequentially (31). Briefly, the inner membrane was solubilized by one hour of incubation at room temperature in 50 mM Tris-HCl, pH 7.5, 1% N-tetradecyl-N,N-dimethyl-3-ammonio-1-propanesulfonate (ZW 3-14), and then the outer membrane by an additional one hour of incubation at room temperature in 50 mM Tris-HCl, pH 7.5-2% ZW 3-14. The periplasmic fraction was prepared from washed intact cells by a cold osmotic shock following the protocol of Osborn and Munson (29). Proteins present in the various samples were analyzed by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining or immunodetection with anti-HasR and anti-MBP antibodies.
HasR N-terminal amino acid sequencing.
The outer membrane proteins of Escherichia coli C600(pBAD24-hasR) were separated by SDS-PAGE and blotted onto an Immobilon membrane. The membrane was stained with Ponceau red to localize HasR and used for N-terminal sequencing by the Service de Microséquence de l'Institut Pasteur.
Electrophoresis and immunological techniques.
Proteins were analyzed by SDS-PAGE followed by Coomassie blue staining. Anti-HasR and anti-HasA rabbit polyclonal sera were used for immunodetection at dilutions of 1/3,000 and 1/5,000, respectively. Anti-MBP antibodies were used at a dilution of 1/5,000.
Dot blot HasA binding assay.
C600 strains carrying the various plasmids were grown in LB medium containing 0.01% arabinose at 37°C and harvested when they reached an OD of 1 at 600 nm. Cell pellets were suspended in TBS (50 mM Tris-HCl, pH 7.5, 150 mM NaCl) to an OD600 of 0.4. Aliquots of 50 μl (approximately 107 CFU) were applied to nitrocellulose filters, and dot blot s with serial dilutions of apo- or holo-HasA or no HasA were performed as described (24). Each protein sample was tested three times.
His-HasA construction and purification.
To introduce an Eco47III site at the 5′ end of hasA, the hasA gene was amplified from pSYC134PAM using the oligonucleotides 5′ AGAGAAAGCGCTTTTTCAGTCA 3′ and the 3′-5′ Forward universal primer. The amplified fragment was digested with Eco47III and HindIII and inserted between the SmaI and HindIII sites of pQE32. This fused the sequence encoding the HasA protein lacking its two first amino acids in phase to a pQE32-encoded N terminus containing six histidines, with the gene under the plac promoter. The hybrid gene was sequenced and found to have no undesired alterations. His-HasA was not secreted and was purified from the cytoplasm.
TG1(pHis-HasAPQE32) was grown in 1 liter of LB medium at 37°C. Isopropylthiogalactopyranoside (IPTG) (1 mM) was added when the culture reached zan OD600 of 0.5, and growth was continued for 3 h. Cells were harvested by centrifugation for 20 min at 5,000 × g at 4°C. The cell pellet was resuspended in 40 ml of 100 mM Tris-HCl, pH 7.5, and the cells were broken by one passage through a French press. The preparation was incubated for 15 min at room temperature with 10 μg/ml RNase and DNase. After centrifugation at 20,000 × g for 1 h at 4°C to remove unbroken cells and cell debris, the supernatant was collected for His-HasA purification. Nickel-agarose beads were washed with equilibration buffer (50 mM Tris-HCl, pH 7.5, 10 mM imidazole) and diluted in 2 volumes of the same buffer. They were mixed with the cell supernatant at a ratio of 1 to 5 (vol/vol) of beads to supernatant and incubated for one hour at 4°C on a wheel. The beads were then washed four times in equilibration buffer, and the fusion protein was eluted with 0.5 M imidazole buffer (50 mM Tris-HCl, pH 7.5, 0.5 M imidazole). An aliquot of the elution fraction was analyzed by SDS-PAGE for the presence and purity of His-HasA.
The eluted fraction was dialyzed overnight against 50 mM Tris-HCl, pH 7.5. The protein concentration and heme content were determined as described above for extracellular HasA. His-HasA was functional and bound heme with high affinity. When purified, the protein was already partially (10%, mol/mol) loaded with intracellular heme. Thus, to have a homogeneous preparation, His-HasA was loaded with heme for binding assays.
Binding assays.
Ten μg of His-HasA (1 volume) was mixed with 5 volumes of equilibrated nickel-agarose beads, washed with 50 mM Tris, pH 7.5-0.08% ZW 3-14, and mixed with 4 OD600 equivalents of membrane and soluble fractions from C600 strains carrying the various plasmids. The mixtures were incubated at room temperature for 1 h.
After three washes in 50 mM Tris-HCl, pH 7.5, 10 mM imidazole, 0.08% ZW 3-14, the fusion protein was eluted with 50 mM Tris-HCl, pH 7.5, 0.5 M imidazole, 0.08% ZW 3-14 buffer. The proteins in the unbound and eluted fractions were analyzed by SDS-PAGE and immunodetection with anti-HasR and anti-HasA antibodies.
Heme and hemophore utilization as porphyrin sources.
Cultures of C600 ΔhemA::km carrying various plasmids were grown in LB medium supplemented with 20 μg/ml of δ-aminolevulinic acid and appropriate antibiotics to an OD600 of 1. Aliquots of 100 μl were plated on LB-0.02% arabinose as indicated in the legend to Table 2. All the experiments were repeated at least six times.
TABLE 2.
Growth with heme or hemophore as the sole porphyrin sourcea
| Strain | Growth
|
||||
|---|---|---|---|---|---|
| Heme (μM)
|
Holo-HasA (μM)
|
||||
| 10 | 5 | 1 | 10 | 1 | |
| C600 ΔhemA(pBAD24-hasR) | ++ | ++ | + | +++ | ++ |
| C600 ΔhemA(pBAD24-hasR Δ11-91) | ++ | ++ | + | +++ | ++ |
| C600 ΔhemA(pBAD24-hasR Δ11-192) | + | + | − | − | − |
| C600 ΔhemA(pBAD24-hasR Δ11-192 + pBAD33-hasR 1-242) | + | + | − | − | − |
| C600 ΔhemA-exbB::Tn10(pBAD24-hasR) | − | − | − | − | − |
| C600 ΔhemA-exbB::Tn10(pBAD24-hasR Δ 11-91) | − | − | − | − | − |
| C600 ΔhemA-exbB::Tn10(pBAD24-hasR Δ 11-192) | + | + | − | − | − |
| C600 ΔhemA hemA-exbB::Tn10(pBAD24-hasR Δ11-192 + pBAD33-hasR 1-242) | + | + | − | − | − |
| C600 ΔhemA(pBAD24-hasR Δ11-192 H603A) | − | − | − | − | − |
| C600 ΔhemA-exbB::Tn10(pBAD24-hasR Δ11-192 H603A) | − | − | − | − | − |
Growth was measured around 5-mm wells filled with 50 μl of the indicated solution. −, no growth; +, halo of growth around the well with a radius of 1 mm; ++, halo of growth around the well with a radius of 5 mm; +++, halo of growth around the well with a radius of 10 mm.
Site-directed mutagenesis and plasmid constructions.
All deletions and site-directed mutagenesis were performed using the Quick Change site-directed mutagenesis kit (Stratagene) and verified by sequencing.
The Δ11-91 deletion of hasR has been described (3). The deletions of the plug were verified by sequencing and the Fse1-AatII hasR fragments were inserted into pBAD24-hasR digested with the same enzymes such that the wild-type 1,255-bp Fse1-AatII segment was replaced by the mutated segments carrying the regulatory region and plug deletions and thus encoding the β-barrels.
The plasmids encoding N-proximal HasR fragments (HasR 1-201, HasR 1-210, and HasR 1-242) were constructed by introducing stop codons into pBAD24-hasR at codons 235, 244, and 276, respectively.
Genes encoding plugs or β-barrels were either expressed from pBAD24 or digested with Nsi and HindIII and inserted into pBAD33 linearized with the same restriction enzymes.
To introduce the H603A mutation into pBAD24-hasRΔ 11-192, the Kpn1-BstZ171 fragment of pBAD24-hasR H603A was exchanged with the Kpn1-BstZ171 fragment of pBAD24-hasR Δ11-192. The oligonucleotides used for the constructs are available upon request.
HasR modeling.
The propensity of amino acids to form β-strands was calculated using the PRED-TMBB method based on a hidden markov model (1). This model predicts transmembrane strands and the topology of β-barrel outer membrane proteins of gram-negative bacteria with a score value of 2.914 that is slightly less than the threshold value of 2.965 but in agreement with the fact that HasR is an outer membrane protein.
The three-dimensional model of HasR was constructed in four steps as follows. First, sequence homology searches were carried out in the Protein Data Bank using SMART (32) and BlastP2 (W. Gish, http://BLAST.wustl.edu) algorithms and sequences homologous to HasR were extracted. The Combinatorial Extension (CE) algorithm (20) was used for multiple alignments of three-dimensional template protein structures. This method determines an optimal alignment of multiple protein structures based on a Monte Carlo optimization method.
Then, the Clustalw program and manual alignment were used for target-template alignment (33) Finally, a three-dimensional model of HasR was constructed using the Modeler program for comparative protein structure modeling (25). The input is an alignment of the sequence to be modeled with the template structures, the atomic coordinates of the templates and a script file. Modeler then calculates a model containing all nonhydrogen atoms by satisfaction restraints on bond distances and dihedral angles. The SCWRL 3.0 program (9) was then used for prediction of protein side chain conformations. The 50 models generated were evaluated using energy criteria and the Procheck program using the pdb files of FecA, FepA, FhuA, and BtupB as templates (26).
RESULTS
Model.
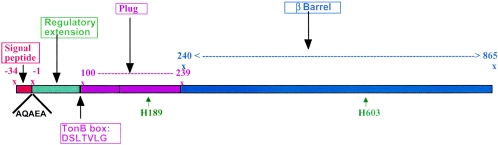
The N-terminal amino acid sequence of the HasR receptor present in the outer membrane preparation of C600(pBAD24-hasR) was determined. The mature protein starts with the sequence AQAE at residue +35 of the coding sequence (Fig. 1). HasR was numbered from the first residue of the mature polypeptide; the signal peptide was given negative numbers (Fig. 1). The crystallographic structures of the FecA, FepA, FhuA, and BtuB receptors were used in the modeling of the HasR structure. A HasR model was constructed from the CE and ClustalW sequence alignment, using Modeler and SCWRL software.
FIG. 1.
HasR domains. HasR numbering starts at the first residue of the mature polypeptide; the signal peptide is given negative numbers. The N-terminal amino acid of the mature protein was determined and is indicated in bold characters above the figure. The regulatory extension encompassing residues 1 to 99 is in green; the plug encompassing residues 100 to 239 is in violet; and the β-barrel, residues 240 to 865, is in blue. The putative TonB box is indicated below the figure. The histidine residue necessary for heme uptake (H603) is indicated below the figure.
Despite an overall low level of sequence identity (FecA/FepA 13.9%, FecA/FhuA 13.5%, FecA/BtuB 16.9%, FepA/FhuA 17.9%, FepA/BtuB 20.3%, and FhuA/BtuB 18.2%), the four transporters display extensive structural similarity: they share a common plug-barrel architecture with similar dimensions; only the extracellular loops and the turns of these transporters diverge in conformation. Despite a very low HasR sequence identity with these receptors (around 10%), we hypothesized that overall structure might be conserved and proposed that HasR consists of three regions, the N-terminal region of HasR (residues 1 to 99), the plug (residues 100 to 239) containing the TonB box motif (residues 100 to 108), and the transmembrane-barrel (residues 240 to 865). A linear representation of the three regions is shown in Fig. 1. The two conserved histidines, H189 and H603, necessary for heme entry through HasR (P. Delepelaire, unpublished), are localized in the plug and in a loop of the barrel, respectively.
Based on this model, we constructed several hasR plug deletions each encoding the signal peptide fused to various parts of the plug followed by the entire β-barrel. We also constructed hasR gene fragments encoding N-terminal segments of the HasR protein of various lengths but excluding the β-barrel.
Production and cellular localization of the mutant proteins.
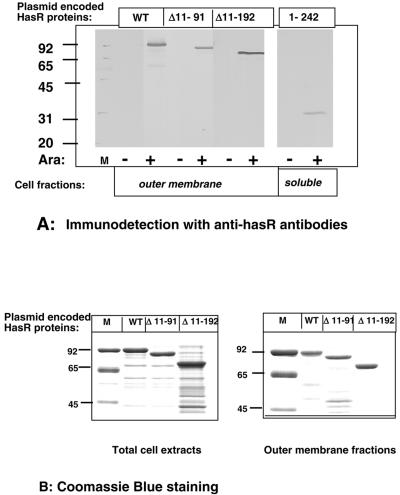
C600 strains harboring pBAD24-hasR encoding the various HasR fragments were grown in LB plus 0.02% arabinose. Cell fractions were analyzed by Western blot with polyclonal anti-HasR antibodies to identify the constructs encoding stable polypeptides. pBAD24-hasR Δ11-91, pBAD24-hasR Δ11-192, and pBAD24-hasR 1-242 encoded stable polypeptides and were used for further analysis. The HasR Δ11-91, HasR Δ11-192, and HasR 1-242 polypeptides were arabinose inducible and were apparently 89 kDa, 79 kDa, and 30 kDa, respectively (Fig. 2A).
FIG. 2.
Production and cellular localization of entire and truncated HasR polypeptides. C600 cells carrying pBAD24-hasR, pBAD24-hasR Δ11-91, pBAD24-hasR Δ11-192, or pBAD24-hasR 1-242 were grown in LB medium with and without 0.02% arabinose at 37°C to an OD600 of 1. Total cell extracts and outer membrane and soluble fractions were prepared. A: Outer membrane and soluble fractions; 0.02 OD600 equivalent of outer membrane fractions or 0.1 OD600 equivalent of soluble fraction was loaded in each lane of an SDS-10% PAGE gel. HasR was immunodetected with a polyclonal anti-HasR serum used at a dilution of 1/5,000. B: Coomassie blue staining of total cell extracts and outer membrane fractions of arabinose-induced cells. Total cellular proteins precipitated with trichloroacetic acid and outer membrane fractons. For total cell extract analysis, 0.2 OD600 equivalent of strains expressing wild-type HasR and HasR Δ11-91 and 0.4 OD600 equivalent of the strain expressing HasR Δ11-192 were loaded. For outer membrane fractions, 1 OD600 equivalent of strains expressing wild-type HasR and HasR Δ11-91 and 2 OD600 equivalent of the strain expressing HasR Δ11-192 were loaded on the gels.
The three HasR polypeptide fragments have a signal peptide and are expected to be exported through the inner membrane. Soluble fractions comprising cytoplasmic and periplasmic compartments and outer membrane fractions were prepared and probed with anti-HasR polyclonal antibodies. The wild-type HasR protein HasR Δ11-91 and HasR Δ11-192 were present in the outer membrane fraction (Fig. 2A). Only traces of immunoreactive material were detected in the soluble fraction (data not shown). HasR 1-242 was in the soluble fraction (Fig. 2A). Extracts of cells producing these three polypeptides did not give cross-reacting material of lower molecular weights on immunoblots, suggesting that the proteins were stable (Fig. 2A). None of these proteins was immunodetected in pellets produced by low-speed centrifugation of cell extracts, indicating that they did not aggregate (data not shown).
Total cell extracts and outer membrane fractions of arabinose-induced cells were studied by SDS-PAGE and Coomassie blue staining: wild-type HasR and HasR Δ11-91 were present in similar amounts in total extracts and in outer membrane fractions. HasR Δ11-192 was also present in both total and outer membrane preparations but was less abundant, about 50% of the wild-type protein level (Fig. 2B). HasR 1-242 was not visible on Coomassie blue-stained gels, suggesting that only very small amounts were produced (data not shown).
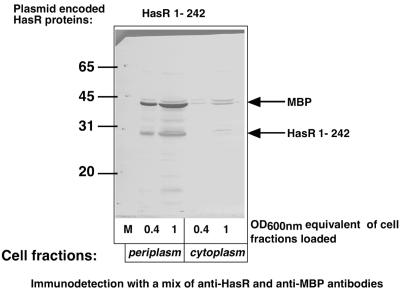
HasR 1-242 was found in the soluble fraction. To determine its localization, C600(pBAD24-hasR 1-242) was grown in LB 0.02% arabinose and 0.4% maltose and was osmotically shocked to release its periplasmic content. HasR 1-242 and the maltose binding protein (MBP), a periplasmic marker, were both mostly detected in the shock fluid (Fig. 3). The small amounts (less than 10%) of MBP detected in the cytoplasmic fraction appear as a doublet, probably precursor and mature forms. Similarly, less than 5% of HasR1-242 immunoreactivity was found in the cytoplasm, and the band exhibited a slightly higher molecular weight than that in the periplasm; the cytoplasm signal therefore presumably corresponds to the precursor form (Fig. 3). Thus, HasR 1-242 is mostly exported to the periplasm.
FIG. 3.
Periplasmic localization of HasR1-242. C600 cells carrying pBAD24-hasR 1-242 were grown in LB medium with 0.02% arabinose and 0.4% maltose at 37°C to an OD600 of 0.75. Periplasmic and cytoplasmic fractions were prepared and the proteins were precipitated with trichloroacetic acid. Aliquots of 0.4 and 1 OD equivalent were loaded in the lanes of an SDS-10% PAGE and proteins were immunodetected with a mix of anti-HasR (1/1,000) and anti-MBP (1/5,000) polyclonal sera.
HasR 1-242 is incorporated into the outer membrane in the presence of HasRΔ11-192.
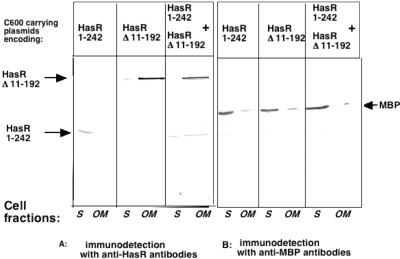
To see whether HasR Δ11-192 and HasR 1-242 interact in vivo, cells expressing both polypeptides were tested for HasR 1-242 incorporation into the outer membrane in the presence of HasR Δ11-192. C600 was transformed with both pBAD33-hasR Δ11-192 and pBAD24-hasR 1-242. Both the plug and the β-barrel polypeptides were produced by this strain. C600(pBAD33-hasR Δ11-192, pBAD24-hasR 1-242), C600(pBAD33-hasR Δ11-192, pBAD24), and C600(pBAD24-hasR 1-242, pBAD33) were grown in LB with 0.02% arabinose and 0.4% maltose. Soluble and outer membrane fractions were prepared, and the presence of HasR 1-242 in both fractions was tested by immunodetection.
The soluble fraction contained both the cytoplasmic and the periplasmic proteins. HasR 1-242 and MBP were mostly periplasmic (Fig. 3), and the soluble fractions are shown in Fig. 4. When expressed alone, HasR 1-242 was in the soluble(periplasmic) fraction and HasR Δ11-192, like the wild-type HasR protein, was in the outer membrane. When coexpressed, HasR 1-242 was found in both fractions, roughly 50% in each (Fig. 4). MBP in the same experiment was mostly in the soluble (periplasmic) fraction. Thus, the plug domain was partially incorporated in the outer membrane fraction in the presence of the β-barrel, strongly suggesting that the two polypeptides interact in the cells.
FIG. 4.
Incorporation of HasR 1-242 into the outer membrane. C600(pBAD33-hasR Δ11-192, pBAD24-hasR 1-242), C600(pBAD33-hasR Δ11-192, pBAD24), and C600(pBAD24-hasR 1-242, pBAD33) cells encoding the indicated polypeptides were grown in LB medium with 0.02% arabinose and 0.4% maltose at 37°C to an OD600 of 1. Outer membrane and soluble fractions were prepared and 0.1 OD600 equivalent of these fractions was loaded in each lane of two SDS-10% PAGE gels. One gel (A) was probed with anti-HasR polyclonal serum (1/1,000) and the other (B) with anti-MBP (1/5,000) polyclonal serum.
Interaction between truncated HasR polypeptides and HasA in vitro.
We tested whether HasR fragments interact with HasA. Outer membrane fractions of cells expressing HasR, HasR Δ11-91, and HasR Δ11-192 and the soluble fraction of cells expressing HasR 1-242 were mixed with His-HasA protein and incubated with Ni-nitrilotriacetic acid (NTA) agarose beads, washed, and eluted from the beads with elution buffer. Unbound and eluted fractions were studied by Western blotting with anti-HasA and anti-HasR polyclonal antibodies.
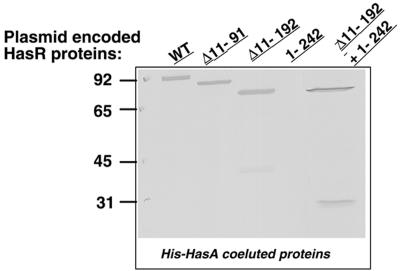
HasR, HasR Δ11-91, and HasR Δ11-192 were retained on the Ni-NTA-agarose only in the presence of His-HasA (data not shown) and were co eluted with His-HasA (Fig. 5). HasR 1-242 was not retained with His-HasA and was present in washes (data not shown). Thus, polypeptides containing the β-barrel interacted with the hemophore, whereas those containing the plug but not the β-barrel did not. The absence of interaction could be the result of inappropriate folding or an insufficient yield during plug preparation. To test this, HasR 1-242 was mixed with HasR Δ11-192 and with His-HasA and applied to Ni-NTA-agarose beads. The three proteins were retained on Ni-NTA-agarose beads and coeluted with His-HasA (Fig. 5). Thus, HasR 1-242 also interacts in vitro with the β-barrel polypeptide. This indicated that the plug protein does not bind to HasA or binds to it with an affinity too low to be detected by affinity copurification.
FIG. 5.
Binding of entire and truncated HasR polypeptides to His-HasA on Ni-NTA. Four OD600 equivalents of outer membrane and cytoplasmic fractions prepared as for Fig. 2A were mixed with Ni-NTA-agarose beads previously loaded with 10 μg of His-HasA. The eluted fractions were loaded onto a SDS-10% PAGE gel and probed with anti-Has-R serum (1/3,000). HasA (not shown in the gel) was present in all the eluted fractions.
Interaction between HasR polypeptides and HasA in vivo.
Solid-phase HasA binding assays were performed to test whether the HasR polypeptides are exposed on the cell surface and interact with HasA.
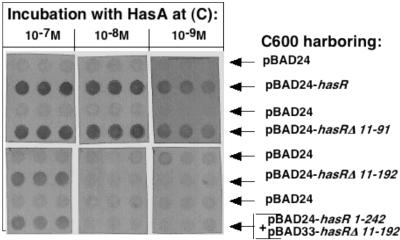
Strains C600(pBAD24), C600(pBAD24-hasR), C600(pBAD24-hasR Δ11-91), and C600(pBAD24-hasR Δ11-192) were grown in LB with 0.01% arabinose to an OD600 of 1 and 107 CFU of each culture were spotted onto filters. The filters were then incubated with various dilutions of HasA and probed with a polyclonal anti-HasA serum. The control strain C600(pBAD24) did not give any signal, whereas the three strains expressing HasR polypeptides reacted with HasA (Fig. 6). The lowest HasA concentration giving a strong positive signal was 10−9 M, for both the strain expressing the wild-type HasR protein and the strain expressing HasR Δ11-91; thus, the receptor lacking the regulatory extension retained a strong affinity for HasA. The HasR Δ11-192 polypeptide reacted with HasA at a higher concentration (10−7 M, Fig. 6). Thus, the β-barrel alone is exposed at the cell surface and interacts with HasA, albeit less efficiently than the native protein. There was no difference between apo- and holo-HasA binding to truncated HasR polypeptides, as is the case for the wild-type HasR protein (data not shown).
FIG. 6.
Solid-phase HasA binding assays. Rows of dots of 107 CFU of C600 cells grown in LB medium with 0.01% arabinose carrying either pBAD24 as a negative control or pBAD24-hasR or pBAD24-hasR Δ11-91or pBAD24-hasR Δ11-192 or pBAD24-hasR 1-242 +pBAD33-hasR Δ11-192 encoding the HasR polypeptides indicated on the figure were incubated with apo-HasA at the concentration indicated above the rows. Each culture was tested in triplicate. The dot blots were probed with polyclonal anti-HasA serum (1/2,000 dilution).
As the plug colocalized to the outer membrane when coexpressed with the β-barrel (Fig. 3), we tested whether coexpression improves the interaction with HasA. Strain C600(pBAD33-hasR Δ11-192, pBAD24-hasR 1-242) was tested for HasA binding as described above. The coexpression of the plug with the barrel did not enhance HasA binding to the β-barrel-expressing cells (Fig. 6).
Phenotypic characterization of the mutant proteins HasR Δ11-91 and HasR Δ11-192.
Strain C600 Δ hemA::km, a heme auxotroph, was transformed with pBAD24-hasR, pBAD24-hasR Δ 11-91 and pBAD24-hasR Δ 11-192 and tested for the ability to use exogenous heme, or holo-hemophore and thus to bypass the heme synthesis mutation and grow aerobically. The strains were plated on LB with 0.02% arabinose and 0.2 mM 2,2′-dipyridyl to induce the TonB complex. Heme or holo-HasA at concentrations from 1 μM to 10 μM was placed in wells in the agar. Strains expressing the wild-type protein and the polypeptide lacking the regulatory extension could use heme bound to the hemophore (as the best heme source). The strain expressing the β-barrel could not use hemophore-bound heme (Table 2). All the strains used heme, although the strain producing the HasR Δ11-192 polypeptide required a fivefold higher heme concentration than the other strains for similar growth (Table 2).
We tested whether coexpression of independently produced plug and β-barrel domains enhanced receptor activity, as was the case for the E. coli ferrichrome (FhuA) and enterobactin (FepA) receptors. Coexpression of the plug and the β-barrel in strain C600 Δ hemA::km(pBAD24-hasR Δ11-192, pBAD33-hasR 1-242) did not improve uptake of either free heme or hemophore-bound heme (Table 2). To determine whether heme uptake involved active transport, which requires the entire TonB-ExbB-ExbD complex, an exbB mutation was introduced into C600 Δ hemA::km carrying the various plasmids. Growth stimulation was ExbB dependent for the wild type and the HasR protein lacking the regulatory extension, but it was unaffected in the strain producing the β-barrel. Thus, heme uptake via the β-barrel was presumably by passive diffusion through the outer membrane. Strain C600 Δ hemA::km(pBAD24-hasR Δ 11-192, pBAD33-hasR 1-242) coexpressing the β-barrel and the plug also displayed only passive heme diffusion (Table 2). Thus, fully active HasR receptor could not be reconstituted from these separately produced domains.
HasR Δ11-192 does not increase nonspecific diffusion through the outer membrane.
Heme uptake by strains expressing HasR Δ11-192 was TonB independent and could be a consequence of nonspecific leakage through the outer membrane. Indeed, HasR Δ11-192 may form nonspecific pores. Such pores would allow the entry of, for example, antibiotics too large to diffuse through porin channels and may disrupt the integrity of the outer membrane, increasing the cells' susceptibility to detergents like SDS. The MICs of vancomycin and SDS for C600 ΔhemA::km, C600 ΔhemA::km(pBAD24-hasR), C600 ΔhemA::km(pBAD24-hasR Δ11-192), and C600 ΔhemA::km(pHSG576 fhuAΔ5-160) were measured. pHSG576 fhuAΔ5-160 encodes the FhuA β-barrel which enhances sensitivity to large antibiotics (5). Neither pBAD24-hasR nor pBAD24-hasR Δ11-192 enhanced the sensitivity of strain C600 ΔhemA::km to SDS or vancomycin. In contrast, pHSG576 fhuAΔ5-160 greatly increased the sensitivity of C600 ΔhemA::km to vancomycin but not to SDS, as previously reported (5). Thus, HasR Δ11-192 neither forms a nonspecific channel nor substantially disturbs outer membrane integrity.
HasR Δ11-192 forms a heme-specific pore in the outer membrane.
The entry of heme through the HasR Δ11-192 pore could be the result of specific heme diffusion. Transport of protoporphyrin IX, the last heme precursor, was tested. None of the strains could use protoporphyrin IX, confirming the specificity for heme. Two conserved histidines are necessary for heme entry through HasR (P. Delepelaire, unpublished). H189 is in the plug, and H603 is in the β-barrel. A mutation of H603A was introduced into HasR Δ11-192 (Materials and Methods). C600 Δ hemA::km(pBAD24-hasR Δ 11-192 H603A) was tested for HasR production, and the H603A mutation did not affect HasR Δ11-192 production (data not shown). The strain was tested for heme acquisition and was unable to use heme (Table 2). Thus, the H603 residue is required for heme uptake through the β-barrel.
HasR 1-242 activity in vivo.
The plug domain interacted with the β-barrel in vivo and in vitro but not in such a way as to reconstitute a fully active receptor when the two were coproduced in vivo. The plug domain is also predicted to interact with TonB. We thus tested whether HasR 1-242 had this activity in vivo.
Strain C600 Δ hemA::km att λ::hasR carrying hasR as a single gene copy on the chromosome was transformed with empty vector pBAD24 or with pBAD24-hasR 1-242 and tested for acquisition of both free and hemophore-bound heme. The strains were plated on LB with 0.02% arabinose and 0.2 mM 2,2′-dipyridyl to induce the TonB complex. Heme acquisition was unaffected by the overproduction of the HasR plug, but heme-hemophore uptake was entirely inhibited (Table 3). Heme acquisition from hemophore requires a higher concentration of the TonB complex than does free heme uptake and also requires more energy (22). Thus, the inhibition of heme-hemophore uptake by the overproduced HasR 1-242 polypeptide could be a consequence of partial TonB trapping. To test this, pNC1, which overproduces the three proteins of the TonB complex, was introduced into strain C600 Δ hemA::km att λ::hasR(pBAD24-hasR1-242). It restored heme-hemophore uptake (Table 3). Thus, inhibition resulted from TonB trapping by the plug. It does not lead to the total loss of TonB-dependent receptor function but affects the function with the highest energy requirement. The TonB binding activity of the plug in vivo strongly suggests that it is in the periplasm and in an active (or at least partially active) conformation.
TABLE 3.
Effect of HasR 1-242 on growth
| Strain | Growth
|
|||
|---|---|---|---|---|
| Heme (μM)
|
Holo-HasA (μM)
|
|||
| 10 | 1 | 10 | 1 | |
| C600 ΔhemA-att λ::hasR(pBAD24, pACYC184) | ++ | + | +++ | ++ |
| C600 ΔhemA-att λ::hasR(pBAD24-hasR 1-242, pACYC184) | ++ | + | − | − |
| C600 ΔhemA-att λ::hasR(pBAD24 hasR 1-242 + pNC1) | ++ | + | +++ | ++ |
Growth was measured around 5-mm wells filled with 50 μl of the indicated solution. −, no growth; +, halo of growth around the well with a radius of 1 mm; ++, halo of growth around the well with a radius of 5 mm; +++, halo of growth around the well with a radius of 10 mm.
DISCUSSION
We used a structural model of HasR for designing constructs made up of various parts of the receptor plug and β-barrel domains linked to a signal peptide. Among the various polypeptides constructed, we used those which were the most stable: the β-barrel—HasR Δ11-192 and the plug—HasR 1-242. The receptor lacking the regulatory domain, HasR Δ11-91, was constructed in previous work. None of these polypeptides formed aggregates. HasR Δ11-91 and HasR Δ11-192 were produced in amounts comparable to that of the wild-type HasR protein. HasR Δ11-91 binds holo-hemophore, allows free and hemophore-bound heme uptake, but is unable to trigger has operon induction (3). Thus, as in the case of the ferric citrate receptor FecA, the regulatory extension is a domain dedicated to regulation and not to transport (14).
HasR Δ11-192 was present in amounts 50% lower than the wild-type protein and was incorporated into the outer membrane and exposed on the cell surface. Dot blotting showed that it interacted with externally added HasA. The HasR 1-242 polypeptide was transported through the inner membrane and was found in the periplasm. Coproduction of both HasR 1-242 and HasR Δ11-192 (the β-barrel) led to partial plug incorporation into the outer membrane, suggesting that the two polypeptides interact in vivo. It is not known where the interaction occurs, after β-barrel incorporation into the outer membrane or in the periplasm with a periplasmic, partially folded β-barrel intermediate. Such intermediates have been described for several outer membrane porins (13, 15).
The HasR plug also interacted with the β-barrel in vitro. HasR Δ11-192, the β-barrel, bound to His-HasA and was copurified with His-HasA on Ni-NTA-agarose, but HasR 1-242 did not bind to His-HasA. When the plug and the β-barrel were added together to His-HasA, both proteins copurified with His-HasA on Ni-NTA-agarose. Ligand binding to a plug in vitro has previously been shown for the enterobactin and FepA plug (34), although the affinity was low. No such interaction was found between transferrin and the TbpA plug (28).
The β-barrel alone also bound HasA in vivo albeit less efficiently than the entire receptor. The difference in HasA binding capacity for wild-type protein and β-barrel alone cannot be imputed to lower β-barrel abundance in the outer membrane, as a strain expressing half of the wild-type protein level binds HasA better than the β-barrel alone (data not shown).
Coexpression of HasR 1-242 and HasR Δ11-192, which colocalize in the outer membrane in vivo, did not enhance HasA binding. This suggests that the hemophore binding domains are carried by the β-barrel and not the plug, unlike heme and siderophores, which bind to residues located in both the β-barrel and the plug (4, 18), (7). However, it is possible that there are additional hemophore binding sites on the plug which were not properly accessible on the reconstituted receptor.
HasR Δ11-192 did not constitute an active TonB-dependent receptor. Coproduction of the plug and β-barrel did not reconstitute an active receptor for free or hemophore-bound heme utilization. In the case of FhuA, such functional complementation has been described for separately encoded plug and β-barrel constructs. The reconstituted FhuA receptor was not entirely functional but nevertheless was able to transport ferrichrome at 45% of the rate of the wild-type FhuA protein and restored wild-type sensitivity to phages and most colicins using FhuA as receptor (5). The negative result with HasR might result from impaired interactions between the two constructs used, which were slightly larger than the plug and β-barrel domains deduced from the structural model. However, other plug and β-barrel constructs with different lengths were not sufficiently stable to be tested.
The plug inhibited heme-hemophore uptake by the wild-type HasR receptor. This inhibition was relieved by overexpression of the TonB complex, indicating that the plug in vivo blocked TonB activity. Most likely, the plug TonB box interacts with TonB as observed in other cases (2). These two observations strongly suggest that HasR 1-242 is at least partially functional. Overproduction of HasR 1-242 did not block other TonB-dependent functions, such as HasR-mediated free heme uptake, which requires lower TonB complex concentrations (22).
An E. coli strain producing the HasR Δ11-192 β-barrel alone was not more sensitive than the wild type to SDS, indicating that β-barrel incorporation into the outer membrane does not seriously perturb membrane integrity. The strain was also no more susceptible than the parental wild-type E. coli or a strain expressing the entire HasR receptor to vancomycin (an antibiotic too large to diffuse through porins). This indicates that HasR Δ11-192 does not form a large open channel in the outer membrane in the absence of the plug to close it. Similar experiments with the FhuA β-barrel domain showed only a slightly increased sensitivity to large antibiotics (5). Indeed, the FhuA β-barrel polypeptide does not form stable channels in lipid bilayers (6). Thus, whereas the channel diameter calculated from the three-dimensional structure of FhuA suggests that the β-barrels might form large open channels in the absence of the plug, this is not observed experimentally either with FhuA (27) or with HasR. The channels might be closed by external loops, or the β-barrels might adopt a different conformation or might collapse when produced without their plugs.
Nevertheless, strains producing the HasR Δ11-192 β-barrel alone were able to use exogenous heme. This transport was not abolished by a mutation in exbB, which inactivates the TonB complex and which was previously shown to abolish active heme transport through entire HasR (30). Thus, the transport was not energy dependent, suggesting that the heme was passively transported. A mutation of histidine 603, which is required for heme uptake through the wild-type receptor, abolished heme transport in strains expressing HasR Δ11-192, indicating that the HasR Δ11-192 channel is heme specific.
Specific channels are usually small β-barrel proteins forming trimers in the outer membrane and allowing energy-independent diffusion of compounds: examples include LamB for maltose and maltodextrins, Tsx for nucleosides, and PhoE for polyphosphates. One such outer membrane protein, Pap31 from Bartonella henselae, binds hemin and allows heme entry when expressed in an E. coli heme auxotroph (38).
Thus, the heme-specific porin that we artificially constructed by splitting HasR into parts might correspond to naturally occurring systems to assimilate heme. We are currently looking for such heme-specific porins.
REFERENCES
- 1.Bagos, P. G., T. Liakopoulos, I. C. Spyropoulos, and S. Hamodrakas. 2004. PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res. (Web Server issue) 32:W400-W404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, P. E., C. D. Nau, J. T. Brown, J. Konisky, and R. J. Kadner. 1990. Genetic suppression demonstrates interaction of TonB protein with outer membrane transport proteins in Escherichia coli. J. Bacteriol. 172:6387-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biville, F., H. Cwerman, S. Létoffé, S. Rossi, V. Drouet, J. M. Ghigo, and C. Wandersman. 2004. Hemophore-mediated signaling in Serratia marcescens: a new mode of regulation for an extra cytoplasmic function (ECF) sigma factor involved in heme-acquisition. Mol. Microbiol. 53:1267-1277. [DOI] [PubMed] [Google Scholar]
- 4.Bracken, C. S., M. T. Baer, A. Abdur-Rashid, W. Helms, and I. Stojiljkovic. 1999. Use of heme-protein complexes by the Yersinia enterocolitica HemR receptor: histidine residues are essential for receptor function. J. Bacteriol. 181:6063-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun, M., F. Endriss, H. Killmann, and V. Braun. 2003. In vivo reconstitution of the FhuA transport protein of Escherichia coli K-12. J. Bacteriol. 185:5508-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun, M., H. Killmann, E. Maier, R. Benz, and V. Braun. 2002. Diffusion through channel derivatives of the Escherichia coli FhuA transport protein. Eur. J. Biochem. 269:4948-4959. [DOI] [PubMed] [Google Scholar]
- 7.Buchanan, S. K., B. S. Smith, L. Venkatramani, D. Xia, L. Esser, M. Palnitkar, R. Chakraborty, D. van der Helm, and J. Deisenhofer. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56-63. [DOI] [PubMed] [Google Scholar]
- 8.Cadieux, N., and R. J. Kadner. 1999. Site-directed disulfide bonding reveals an interaction site between energy-coupling protein TonB and BtuB, the outer membrane cobalamin transporter. Proc. Natl. Acad. Sci. USA 96:10673-10678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canutescu, A., A. Shelenkov, and R. J. Dunbrack. 2003. A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 12:2001-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chimento, D. P., A. K. Mohanty, R. J. Kadner, and M. C. Wiener. 2003. Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat. Struct. Biol. 10:394-401. [DOI] [PubMed] [Google Scholar]
- 11.Cobessi, D., H. Celia, N. Folschweiller, I. J. Schalk, M. A. Abdallah, and F. Pattus. 2005. The crystal structure of the pyoverdine outer membrane receptor FpvA from Pseudomonas aeruginosa at 3.6A resolution. J. Mol. Biol. 347:121-134. [DOI] [PubMed] [Google Scholar]
- 12.Cobessi, D., H. Celia, and F. Pattus. 2004. Crystallization and X-ray diffraction analyses of the outer membrane pyochelin receptor FptA from Pseudomonas aeruginosa. Acta Crystallogr. D Biol. Crystallogr. 60:1919-1921. [DOI] [PubMed] [Google Scholar]
- 13.Duguay, A. R., and T. J. Silhavy. 2002. Signal sequence mutations as tools for the characterization of LamB folding intermediates. J. Bacteriol. 184:6918-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enz, S., S. Mahren, U. H. Stroeher, and V. Braun. 2000. Surface signaling in ferric citrate transport gene induction: interaction of the FecA, FecR, and FecI regulatory proteins. J. Bacteriol. 182:637-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eppens, E. F., N. Nouwen, and J. Tommassen. 1997. Folding of a bacterial outer membrane protein during passage through the periplasm. EMBO J. 16:4295-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferguson, A. D., R. Chakraborty, B. S. Smith, L. Esser, D. van der Helm, and J. Deisenhofer. 2002. Structural basis of gating by the outer membrane transporter FecA. Science 295:1715-1719. [DOI] [PubMed] [Google Scholar]
- 17.Ferguson, A. D., and J. Deisenhofer. 2004. Metal import through microbial membranes. Cell 116:15-24. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson, A. D., E. Hofmann, J. W. Coulton, K. Diederichs, and W. Welte. 1998. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282:2215-2220. [DOI] [PubMed] [Google Scholar]
- 19.Ghigo, J. M., S. Létoffé, and C. Wandersman. 1997. A new type of hemophore-dependent heme acquisition system of Serratia marcescens reconstituted in Escherichia coli. J. Bacteriol. 179:3572-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guda, C., E. D. Scheeff, P. E. Bourne, and I. N. Shyndyalov. 2001. A new algorithm for the alignment of multiple protein structures using Monte Carlo optimization. Pacific Symp. Biocomput. 6:275-286. [DOI] [PubMed] [Google Scholar]
- 21.Létoffé, S., L. Debarbieux, N. Izadi, N. Delepelaire, and C. Wandersman. 2003. Ligand delivery by heme carrier proteins: the binding of Serratia marcescens hemophore to its outer membrane receptor is mediated by two distinct peptide regions. Mol. Microbiol. 50:77-88. [DOI] [PubMed] [Google Scholar]
- 22.Létoffé, S., P. Delepelaire, and C. Wandersman. 2004. Free and hemophore-bound heme acquisitions through the outer membrane receptor HasR have different requirements for the TonB-ExbB-ExbD complex. J. Bacteriol. 186:4067-4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Létoffé, S., J. M. Ghigo, and C. Wandersman. 1994. Secretion of the Serratia marcescens HasA protein by an ABC transporter. J. Bacteriol. 176:5372-5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Létoffé, S., F. Nato, M. E. Goldberg, and C. Wandersman. 1999. Interactions of HasA, a bacterial haemophore, with haemoglobin and with its outer membrane receptor HasR. Mol. Microbiol. 33:546-555. [DOI] [PubMed] [Google Scholar]
- 25.Marti-Renom, M. A., A. C. Stuart, A. Fiser, R. Sanchez, F. Melo, and A. Sali. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29:291-325. [DOI] [PubMed] [Google Scholar]
- 26.Morris, A. L., M. W. MacArthur, E. G. Hutchinson, and J. M. Thornton. 1992. Stereochemical quality of protein structure coordinates. Proteins 12:345-364. [DOI] [PubMed] [Google Scholar]
- 27.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oke, M., R. Sarra, R. Ghirlando, S. Farnaud, A. Gorringe, R. Evans, and S. Buchanan. 2004. The plug domain of a neisserial TonB-dependent transporter retains structural integrity in the absence of its transmembrane beta-barrel. FEBS Lett. 564:294-300. [DOI] [PubMed] [Google Scholar]
- 29.Osborn, M. J., and R. Munson. 1974. Separation of the inner (cytoplasmic) and outer membranes of Gram-negative bacteria. Methods Enzymol. 31:642-653. [DOI] [PubMed] [Google Scholar]
- 30.Paquelin, A., J. M. Ghigo, S. Bertin, and C. Wandersman. 2001. Characterization of HasB, a Serratia marcescens TonB-like protein specifically involved in the haemophore-dependent haem acquisition system. Mol. Microbiol. 42:995-1005. [DOI] [PubMed] [Google Scholar]
- 31.Schalk, I. J., P. Kyslik, D. Prome, A. van Dorsselaer, K. Poole, M. A. Abdallah, and F. Pattus. 1999. Copurification of the FpvA ferric pyoverdin receptor of Pseudomonas aeruginosa with its iron-free ligand: implications for siderophore-mediated iron transport. Biochemistry 38:9357-9365. [DOI] [PubMed] [Google Scholar]
- 32.Schultz, J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95:5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Usher, K. C., E. Ozkan, K. H. Gardner, and J. Deisenhofer. 2001. The plug domain of FepA, a TonB-dependent transport protein from Escherichia coli, binds its siderophore in the absence of the transmembrane barrel domain. Proc. Natl. Acad. Sci. USA 98:10676-10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vakharia, H., and K. Postle. 2002. FepA with globular domain deletions lacks activity. J. Bacteriol. 184:5508-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wandersman, C., and P. Delepelaire. 2004. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 58:611-647. [DOI] [PubMed] [Google Scholar]
- 37.Wandersman, C., and I. Stojiljkovic. 2000. Bacterial heme sources: role of hemophores and receptors. Curr. Opin. Microbiol. 3:215-220. [DOI] [PubMed] [Google Scholar]
- 38.Zimmermann, R., V. Kempf, E. Schiltz, K. Oberle, and A. Sander. 2003. Hemin binding, functional expression, and complementation analysis of Pap 31 from Bartonella henselae. J. Bacteriol. 185:1739-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]