Figure 3. Regulatory networks mediating splicing neoantigen burden in Melanoma.
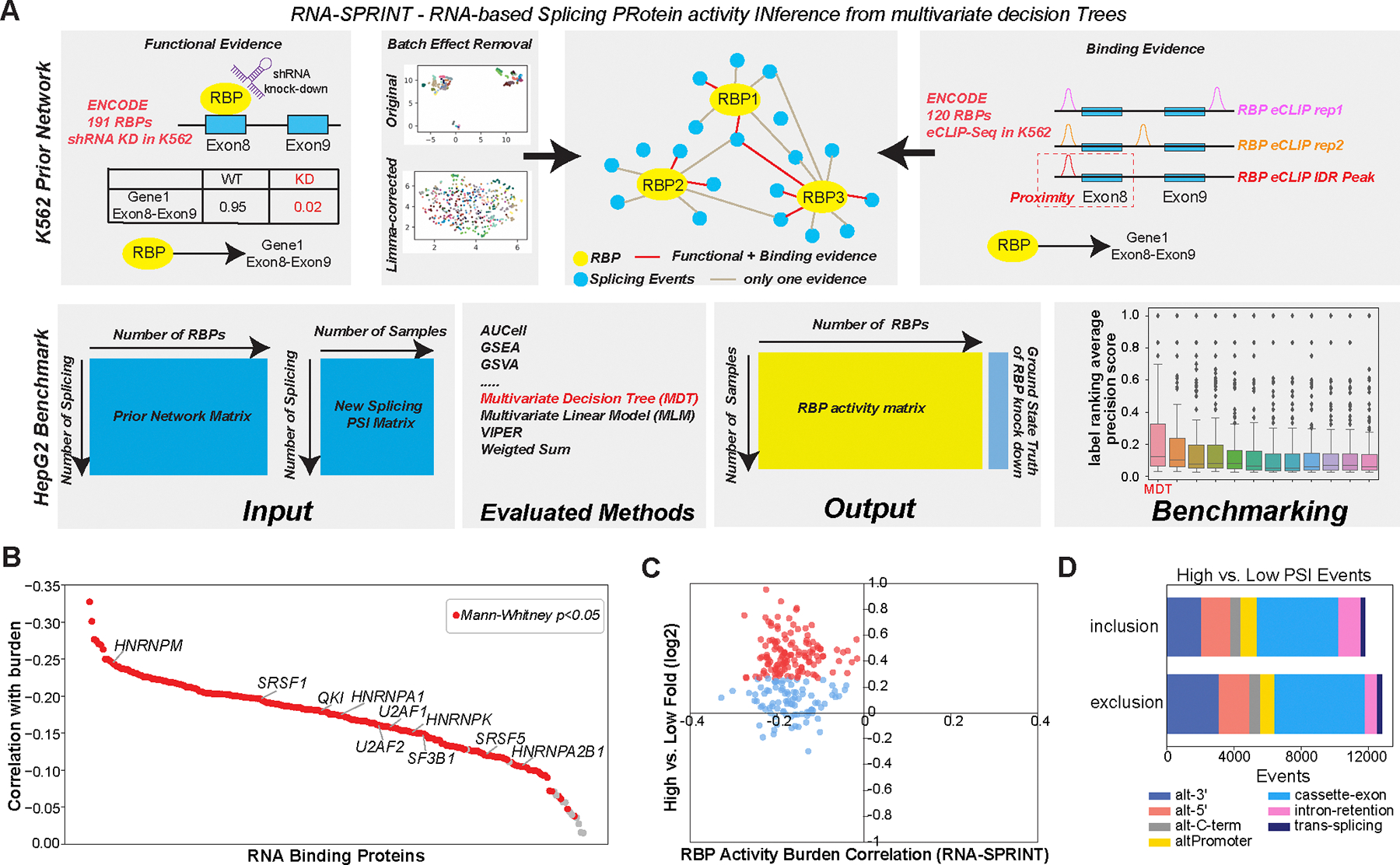
A) Schematic overview of the software RNA-SPRINT and associated benchmarking steps. The workflow involves construction of an RNA Binding Protein (RBP) prior network to predict splicing regulatory interactions. Evaluation of the method is overviewed, consisting of RNA-SPRINT benchmarking relative to 12 transcription factor (TF) activity methods in HepG2 cell line RBP knockdown RNA-Seq datasets. B) The correlation of inferred RBP activity with splicing neoantigen burden for all TCGA patients with melanoma. C) Comparison of RBP activity-burden correlations with RBP differential gene expression, for high versus low burden (TCGA SKCM). Red = upregulated genes (fold>1.2 and eBayes t-test p<0.05, FDR corrected) in high burden. D) Type of splicing events observed with exon/intron inclusion or exclusion comparing high versus low burden.
