Figure 4. Shared splicing neoantigens are frequently detected by MS and are defined by their sequence composition in Melanoma.
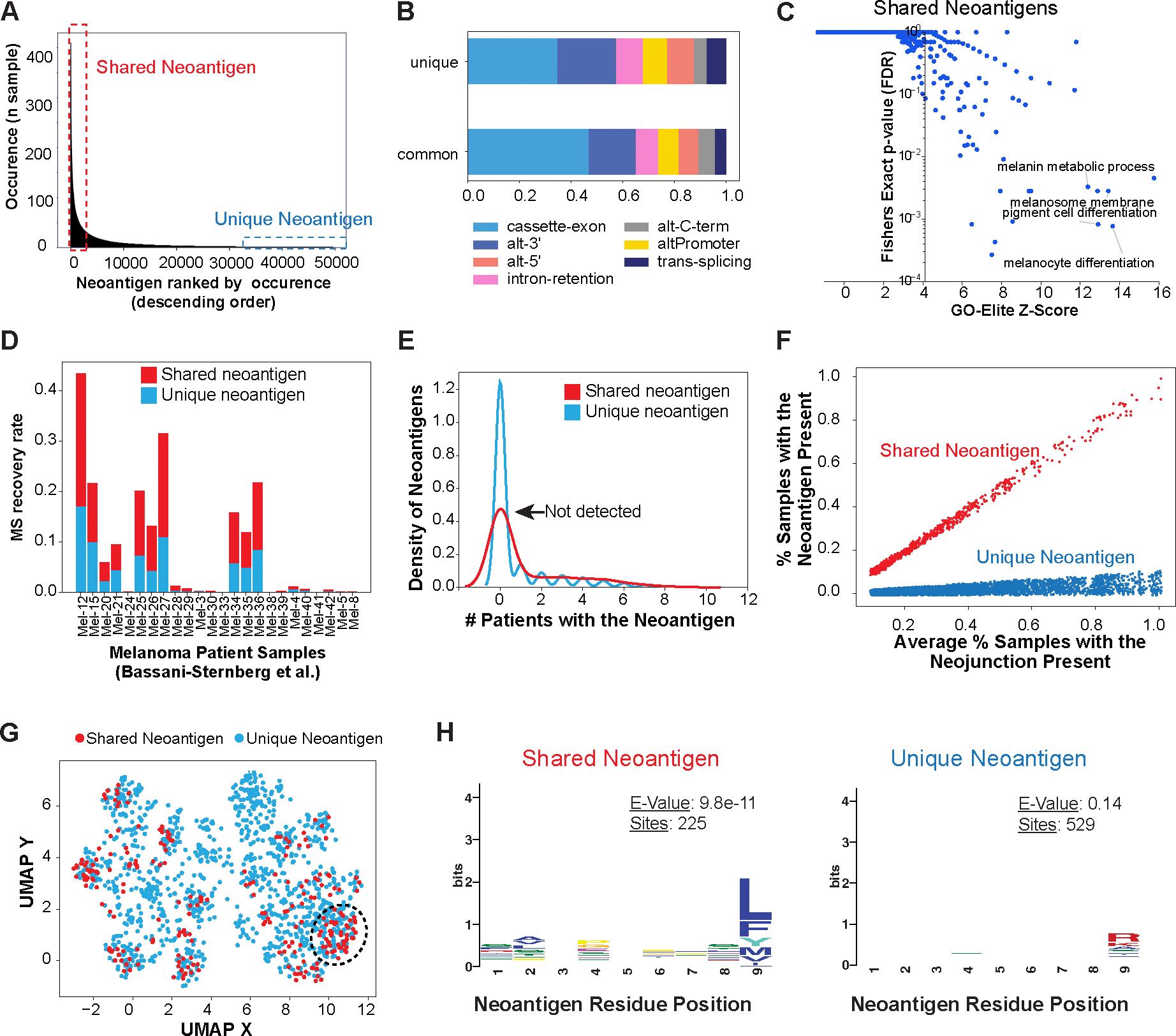
A) Identification of common (shared) and unique immunogenic splicing neoantigens in the TCGA Melanoma cohort, based on their frequency of occurrence among patients. B) Frequency of splicing-event types for shared and unique splicing neoantigen junctions in TCGA. C) Gene-set enrichment with GO-Elite of the Gene Ontology and pathways of shared neoantigens (present in >15% of patients with melanoma). D) MS recovery rate in an independent melanoma immunopeptidome dataset (Bassani-Sternberg et al.) between shared and unique neoantigens considered in the query database. E) Kernel density estimate plot comparing the observed occurrence in an independent immunopeptidomics MS experimental cohort, for all detected shared (>15% of patients with melanoma) versus unique splicing-neoantigens. F) Re-defined shared and unique neoantigens in TCGA by normalizing the occurrence of their parental splice junction, leveraging their respective observed amino acid bias. G) UMAP of splicing neoantigens based on their amino acid physiological properties in TCGA, highlighting neoantigens that cluster based on shared amino acid physicochemical features. H) Distinct enriched amino acid motifs (MEME), comparing shared versus unique neoantigens.
