Figure 7. SNAF-B finds full-length mRNAs and stable-proteoforms for targeted therapies.
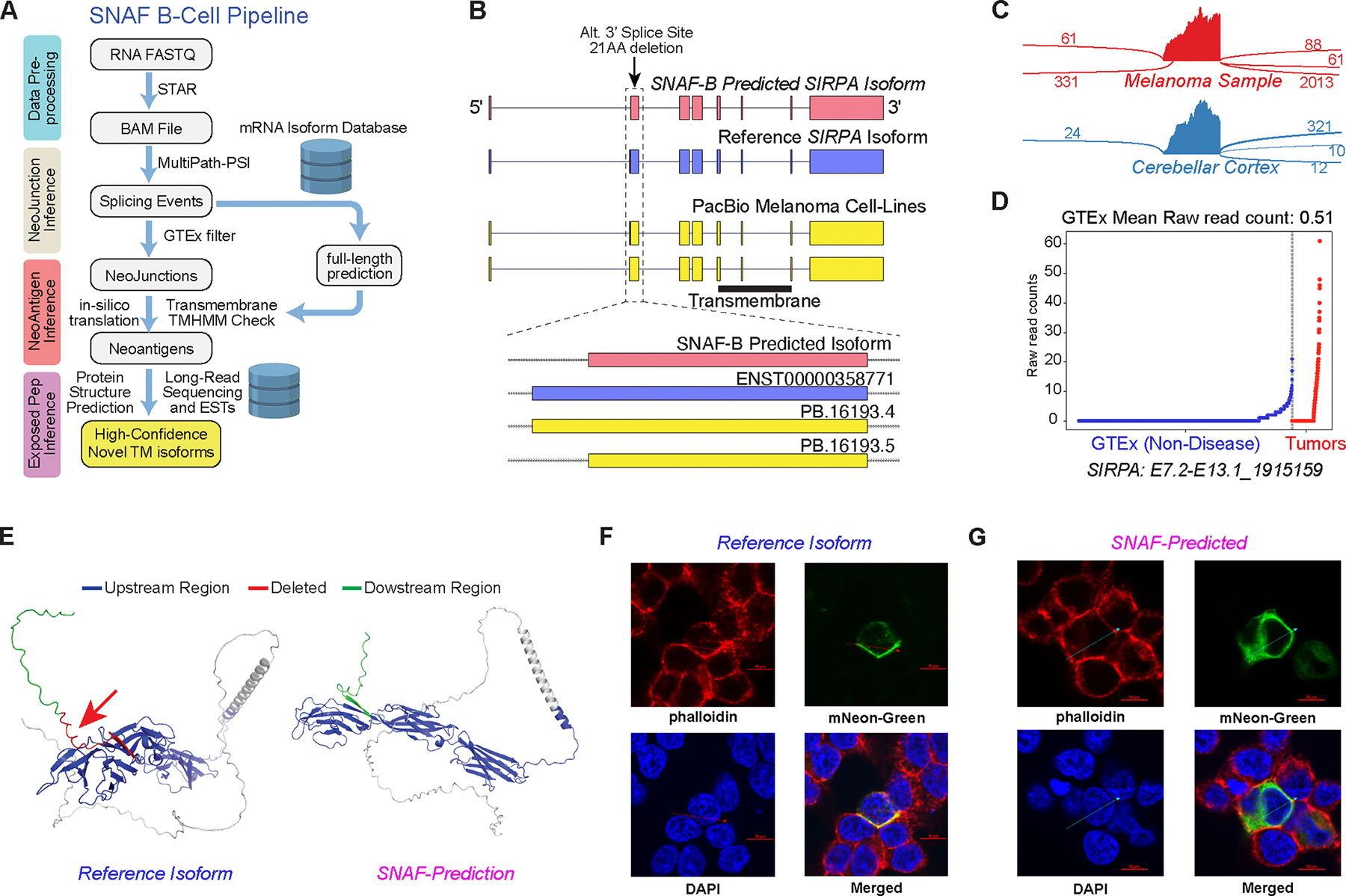
A) Overview of the SNAF-B prediction workflow to define ExNeoEpitopes. The workflow begins with bulk RNA-Seq datasets and optional long-read sequencing data integration to produce results with multiple levels of in silico evidence. B) Comparison of a SNAF-B predicted full-length isoform in the transmembrane protein SIRPA to documented mRNA isoforms and those predicted from PacBio long-read IsoSeq of melanoma cell lines. C) SashimiPlot of alternative 3’ splice site selection in Melanoma and Brain RNA-Seq for SIRPA. D) Specificity of the indicated SIRPA ExNeoEpitope for TCGA melanoma samples versus an integrated healthy controls tissue database (GTEx + TCGA). E) Alphafold2 3D modeling of the reference isoform and the long-read verified ExNeoEpitope. Arrow denotes the deleted region in the alternative isoform. F,G) Co-localization of the SIRPA reference (F) or Melanoma-specific (G) splice isoform by confocal microscopy with a cell surface stain (phalloidin). The arrow indicates the cross-section used to quantify fluorophore spatial coincidence.
