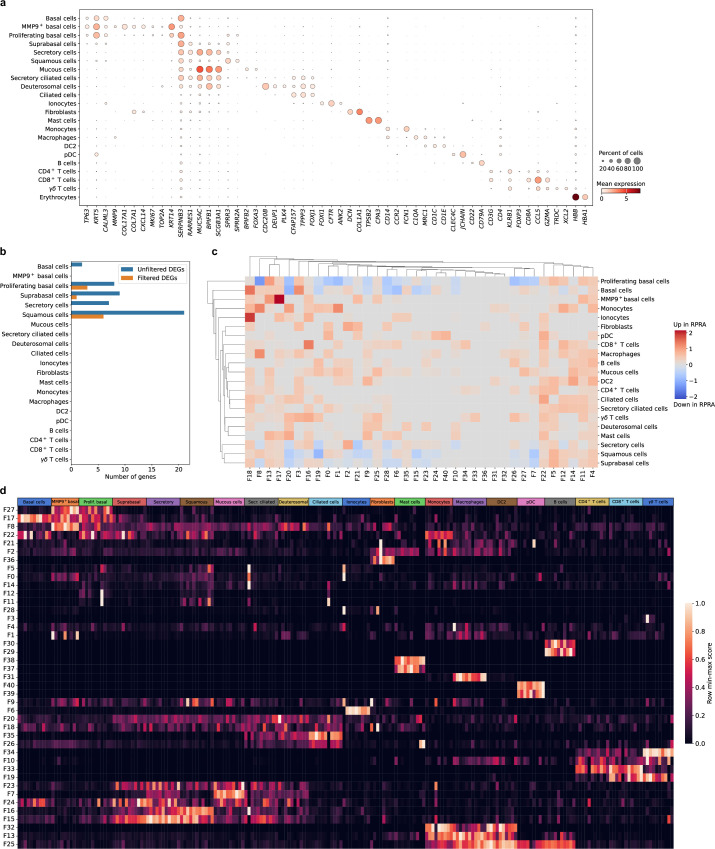
Extended Data Fig. 7. Transcriptomic changes in the nasal mucosa do not reflect ongoing inflammation in the distal lung in patients with RPRA.
a. Dot plot showing expression of the selected cell type markers used to identify cell types in the integrated single-cell RNA-seq data from 5 patients with RPRA and 6 healthy control subjects from Fig. 7a. b. Bar plot showing the number of differentially expressed genes in different cell types between 5 patients with RPRA and 6 healthy control subjects (q < 0.05, Wald test with FDR correction) with and without filtering criteria applied. c. Hierarchical clustering on the signal-to-noise ratio of Spectra subject scores between patients with RPRA and healthy controls. No factors were differentially expressed (q < 0.05, Wilcoxon rank-sum test on subject scores with FDR correction). d. Heatmap of subject scores for Spectra programs. Each column represents a single subject. Rows are min-max scaled. MMP9hi basal cells (MMP9+ basal), proliferating basal cells (Prolif. Basal), suprabasal cells (Suprabasal), secretory cells (Secretory), basal cells (Basal), secretory ciliated cells (Secr. Ciliated), deuterosomal cells (Deuterosomal), type II conventional dendritic cells (DC2), plasmacytoid dendritic cells (pDC).