Abstract
The emergence of ciprofloxacin-resistant Salmonella enterica serovar Choleraesuis in recent years has become an important public health issue in Taiwan. The resistant strains that cause human infections are considered to be from pigs. In this study, we characterized 157 swine and 42 human Salmonella serovar Choleraesuis isolates by pulsed-field gel electrophoresis (PFGE) and drug susceptibility testing to investigate the epidemiologic relationship among the isolates. By PFGE analyses, two major clusters (clusters GA and GB) were identified. Isolates in cluster GA were of both human and swine origins, while those in cluster GB were from pigs only. Among the various genotypes identified, genotype gt-1a was the most prevalent, which was found in 71% (30 of 42) and 48% (76 of 157) of human and swine isolates, respectively. The susceptibility tests for the 106 gt-1a isolates identified 44 susceptibility profiles and showed that 73% of human isolates and 34% of swine isolates were resistant to three fluoroquinolones (ciprofloxacin, enrofloxacin, and norfloxacin). Our findings indicate that a clonal group of Salmonella serovar Choleraesuis may have been circulating in human and swine populations in Taiwan for years and that the fluoroquinolone-resistant Salmonella serovar Choleraesuis strains most likely evolved from a gt-1a clone that emerged in 2000 and that then caused widespread infections in humans and pigs. Nevertheless, it is still debatable whether those Salmonella infections in humans are caused by isolates derived from pigs, on the basis of the higher fluoroquinolone and other antimicrobial resistance percentages in human isolates than in pig isolates.
Salmonella enterica serovar Choleraesuis, including variety Kunzendorf, is a host-adapted serovar in swine since it is able to cause diseases and circulate mainly in pig populations by direct transmission (22). Salmonella serovar Choleraesuis, which primarily causes septicemia in pigs, can be characterized by pneumonia, liver abscesses, and enteritis (7). This pathogen can occasionally infect humans. It often causes severe invasive diseases, primarily septicemia with little involvement of the intestinal tract in humans, especially in elderly people with underlying diseases (8, 12). As a major isolated serovar of Salmonella from swine (13), the impact of Salmonella serovar Choleraesuis on public health differs greatly in different geographic areas. Salmonella serovar Choleraesuis is an infrequent serovar isolated from human sources in the United States, Canada, and the United Kingdom (2, 6, 20). Moreover, it was the 11th most common serovar causing human salmonellosis in Thailand from 1993 to 2002 (1). However, in Taiwan, Salmonella serovar Choleraesuis has become of particular concern since it was the second most common serovar of salmonellas isolated from humans from 1991 to 1996 (9, 10).
Antimicrobial therapy is necessary for the treatment of Salmonella serovar Choleraesuis infections, because approximately 78% of the infected individuals in Taiwan developed bacteremia with little or no intestinal involvement (12). The conventional antimicrobial agents, including ampicillin, chloramphenicol, and trimethoprim-sulfamethoxazole, used to be the drugs of choice for the treatment of salmonellosis before the 1980s. Since strains resistant to these antimicrobial agents have become highly prevalent in recent decades (9, 11), extended-spectrum cephalosporins and fluoroquinolones have been used as alternative agents for the treatment of salmonella infections. Recently, the emergence of fluoroquinolone resistance in Salmonella serovar Choleraesuis has become a threat to the public health in Taiwan. A study conducted in a medical center in northern Taiwan showed that no ciprofloxacin resistance was detected in Salmonella serovar Choleraesuis before 1999, but a dramatic increase in the incidence of ciprofloxacin resistance has been observed since 2000 (11). In 2000 and 2001, the observed annual rate of resistance was more than 40% among isolates collected in the medical center. In contrast, the rate of resistance among isolates collected across Taiwan in 2001 was only 7.5% (17). Based on the molecular identification of common genotypic isolates of human and swine origin as well as the prolonged use of a fluoroquinolone (i.e., enrofloxacin) in pig feed as a growth promoter, the authors of these two studies concluded that the ciprofloxacin-resistant Salmonella serovar Choleraesuis strains were transmitted from pigs to humans (11, 17, 23). However, the transmission direction of ciprofloxacin-resistant strains between humans and swine remains debatable.
In this study, to investigate the epidemiologic relationship of Salmonella serovar Choleraesuis strains from humans and pigs, Salmonella serovar Choleraesuis isolates were characterized by pulsed-field gel electrophoresis (PFGE) genotyping and antimicrobial susceptibility testing. Based on our findings, it is highly debatable whether pigs were the source of ciprofloxacin-resistant strains of Salmonella serovar Choleraesuis strains collected from humans between 1997 and 2002.
MATERIALS AND METHODS
Salmonella serovar Choleraesuis isolates.
A total of 199 Salmonella serovar Choleraesuis isolates (42 from humans and 157 from pigs) were collected from 1997 to 2002. The numbers of isolates recovered from human sources each year from 1997 to 2002 were 5, 0, 2, 9, 14, and 12, respectively; and the numbers of isolates recovered from swine sources each year from 1997 to 2002 were 8, 24, 25, 34, 28, and 38, respectively. Human isolates were kindly provided by several local medical centers in Taiwan, including National Taiwan University Teaching Hospital (5 isolates), Taipei Veterans General Hospital (3 isolates), Kaohsiung Veterans General Hospital (2 isolates), Chi-Mei Medical Center (2 isolates), and Chang Gung Memorial Hospital (21 isolates), and from the Third Branch Office, Center for Disease Control, in Taichung (9 isolates). The swine isolates, mainly recovered from the septicemia cases collected on farms, were offered by Chao-Fu Chang. These clinical cases were from different pig herds located in six counties (Changhwa, Yunlin, Chiayi, Tainan, Kaohsiung, and Pingtung) in south-central Taiwan. For serotype identification, antisera for the detection of O and H antigens were purchased from S&A Reagents Lab Limited (Thailand) and Denka Seiken Co., Ltd. (Japan). All isolates were confirmed to be Salmonella serovar Choleraesuis variety Kunzendorf, according to the Kaufmann-White scheme and the serotyping protocols developed by the Centers for Disease Control and Prevention, Atlanta, Ga. (4).
PFGE analysis.
PFGE analysis was carried out as described previously (14), with minor modifications. The DNA embedded in agarose plugs was digested with 10 U/plug slice of the XbaI restriction enzyme (Promega Corp.), following the manufacturer's recommendations. Restriction fragments were separated by PFGE in 1% SeaKem Gold agarose gels (Unimed Healthcare Inc., Taipei, Taiwan) in 0.5× Tris-borate-EDTA (45 mM Tris borate, 1 mM EDTA, pH 8.3) buffer at 14°C in a CHEF mapper apparatus (Bio-Rad Laboratories, Richmond, Calif.). Electrophoresis conditions were as follows: initial switch time of 2.16 s, final switch time of 54.17 s, 6 V/cm, and an angle of 120° for 19 h. Fragments of the XbaI-digested genomic DNA of S. enterica serovar Braenderup H9812, kindly provided by Bala Swaminathan at the Centers for Disease Control and Prevention, were loaded in the gel as a reference standard marker. After electrophoresis, the PFGE patterns were imaged with a digital camera system at 1,792 by 1,200 pixels.
Analysis of PFGE patterns.
The PFGE gel image was analyzed with BioNumerics software (Applied Maths, Belgium), and the patterns were stored in a Salmonella fingerprint database. The dendrogram, which consisted of the XbaI PFGE patterns, was constructed by the unweighted pair group method with arithmetic means (UPGMA) algorithm, and the Dice similarity of two patterns was determined with the software by the use of 3% optimization and 1% position tolerance. Bacterial isolates with one or more band differences were considered to be of different PFGE genotypes.
Antimicrobial susceptibility testing by disk diffusion method.
Antimicrobial susceptibility tests for Salmonella serovar Choleraesuis were conducted by the standard disk diffusion method according to CLSI (formerly the National Committee for Clinical Laboratory Standards) (24). The antimicrobial agents used included ampicillin (10 μg/disk), cephalothin (30 μg/disk), chloramphenicol (30 μg/disk), ciprofloxacin (5 μg/disk), enrofloxacin (5 μg/disk), nalidixic acid (30 μg/disk), nitrofurantoin (300 μg/disk), norfloxacin (10 μg/disk), gentamicin (10 μg/disk), streptomycin (10 μg/disk), tetracycline (30 μg/disk), and trimethoprim-sulfamethoxazole (1.25-23.75 μg/disk).
Statistical analysis.
Chi-square tests were conducted to test the homogeneity of antimicrobial resistance between groups of isolates by using SPSS, version 10.0. Statistical significance was determined when the P value was less than 0.05.
RESULTS
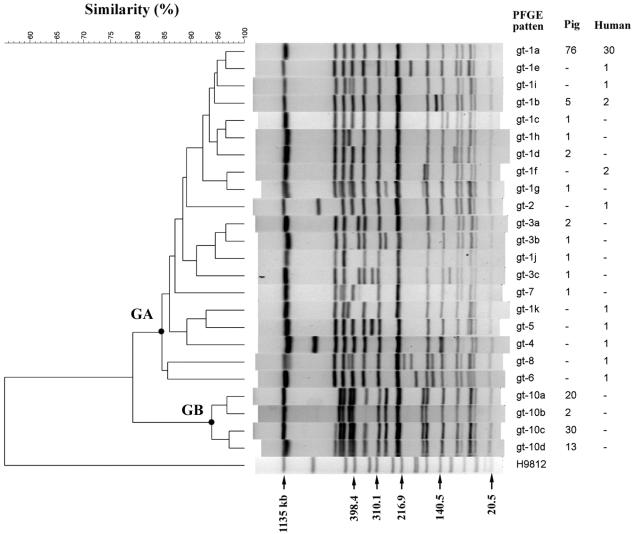
A total of 24 PFGE patterns were identified among the 199 isolates of Salmonella serovar Choleraesuis. The clustering analysis for the PFGE patterns revealed two distinct groups, cluster GA and cluster GB (Fig. 1). Cluster GA consisted of 20 PFGE patterns (genotypes), and isolates from both humans and pigs of the gt-1a and gt-1b genotypes were detected in this cluster. Genotype gt-1a was the most prevalent. It was identified in 30 human and 76 swine isolates, which were collected from various locations in western Taiwan from 1997 to 2002. The phylogenetic and epidemiologic data suggested that the genotypes in cluster GA could have evolved from a common ancestor and that these genotypic Salmonella serovar Choleraesuis isolates had been circulating in human and swine populations for years. Interestingly, no human isolate belonged to cluster GB. Cluster GB consisted of four PFGE patterns, which were found only in 65 pig isolates collected from 1998 to 2002 on pig farms located in Pingtung, the southernmost county in Taiwan.
FIG. 1.
Dendrogram and PFGE patterns of XbaI-digested chromosomal DNA of S. enterica serovar Choleraesuis and the numbers of isolates of pig and human origin. The dendrogram was constructed by use of the UPGMA algorithm and the Dice similarity coefficient by using BioNumerics software with 3% optimization and 1% position tolerance.
Antimicrobial susceptibility tests with 12 antimicrobial agents and the 199 Salmonella serovar Choleraesuis isolates showed that more than 95% of the isolates were resistant to ampicillin, chloramphenicol, nalidixic acid, and tetracycline (Table 1) but that only a few (1%) were resistant to cephalothin. In contrast, 29% of the isolates were resistant to three fluoroquinolones (ciprofloxacin, enrofloxacin, and norfloxacin); and about half of the isolates were resistant to gentamicin, nitrofurantoin, streptomycin, and trimethoprim-sulfamethoxazole. Among all isolates tested, 52% were multidrug resistant (resistant to seven or more of the antimicrobial agents that we tested). The prevalences of multidrug resistance among the isolates from humans and swine were 76% and 46%, respectively. The statistical analysis further revealed that the human isolates had significantly higher rates of resistance than the swine isolates to cephalothin, ciprofloxacin, enrofloxacin, norfloxacin, streptomycin, and trimethoprim-sulfamethoxazole but had lower rates of resistance to chloramphenicol and nitrofurantoin (P < 0.05).
TABLE 1.
Prevalence of resistance to antimicrobial agents among different groups of S. enterica serovar Choleraesuis isolates
| Antimicrobial agent | All isolates (% resistant) (n = 199) | All isolates
|
PFGE cluster
|
PFGE gt-1a
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Human (n = 42) (% resistant) | Swine (n = 157) (% resistant) | P valuea | GA (n = 134) (% resistant) | GB (n = 65) | P valuea | Human (n = 30) (% resistant) | Swine (n = 76) (% resistant) | P valuea | ||
| Ampicillin | 95 | 95 | 96 | 0.933 | 93 | 100 | 0.032 | 97 | 97 | 0.844 |
| Cephalothin | 1 | 5 | 0 | 0.007 | 1 | 0 | 0.463 | 7 | 0 | 0.030 |
| Chloramphenicol | 96 | 86 | 99 | 0.001 | 94 | 100 | 0.132 | 87 | 97 | 0.100 |
| Ciprofloxacin | 29 | 64 | 19 | <0.001 | 43 | 0 | <0.001 | 73 | 34 | <0.001 |
| Enrofloxacin | 29 | 64 | 19 | <0.001 | 43 | 0 | <0.001 | 73 | 34 | <0.001 |
| Gentamicin | 47 | 43 | 48 | 0.852 | 46 | 49 | 0.867 | 40 | 51 | 0.510 |
| Nalidixic acid | 99 | 98 | 100 | 0.053 | 99 | 100 | 0.485 | 97 | 100 | 0.110 |
| Nitrofurantoin | 47 | 17 | 55 | <0.001 | 31 | 80 | <0.001 | 13 | 45 | 0.003 |
| Norfloxacin | 29 | 64 | 19 | <0.001 | 43 | 0 | <0.001 | 73 | 34 | <0.001 |
| Streptomycin | 61 | 88 | 54 | <0.001 | 73 | 37 | <0.001 | 93 | 67 | 0.019 |
| Tetracycline | 97 | 98 | 97 | 0.951 | 96 | 100 | 0.115 | 97 | 99 | 0.492 |
| SxTb | 42 | 86 | 31 | <0.001 | 61 | 3 | <0.001 | 90 | 49 | <0.001 |
By chi-square test.
SxT, trimethoprim-sulfamethoxazole.
To see whether the rates of resistance were different for the two distinct PFGE genotypic clusters, we compared the rates of resistance among the isolates in clusters GA and GB. The results showed significant differences in the rates of resistance to the seven antimicrobial agents tested between these two genotypic groups (Table 1). Of most importance, all of the isolates in cluster GB were susceptible to the three fluoroquinolones tested and trimethoprim-sulfamethoxazole. All isolates in cluster GB had closely related PFGE patterns, and all were of swine origin.
To avoid interference in the statistical analysis for drug resistance by the different genotypes of the isolates, we chose the 106 isolates with the same gt-1a genotype (30 from humans and 76 from pigs) for further comparison. The results showed that the human and swine groups had significant differences in rates of resistance to the seven antimicrobials (Table 1). Compared with the swine isolates, the human isolates still had higher rates of resistance to fluoroquinolones but had lower rates of resistance to nitrofurantoin. Contrary to what was found for the pig isolates, our results further indicated that human isolates with fluoroquinolone resistance were more likely to be susceptible to nitrofurantoin (P < 0.05).
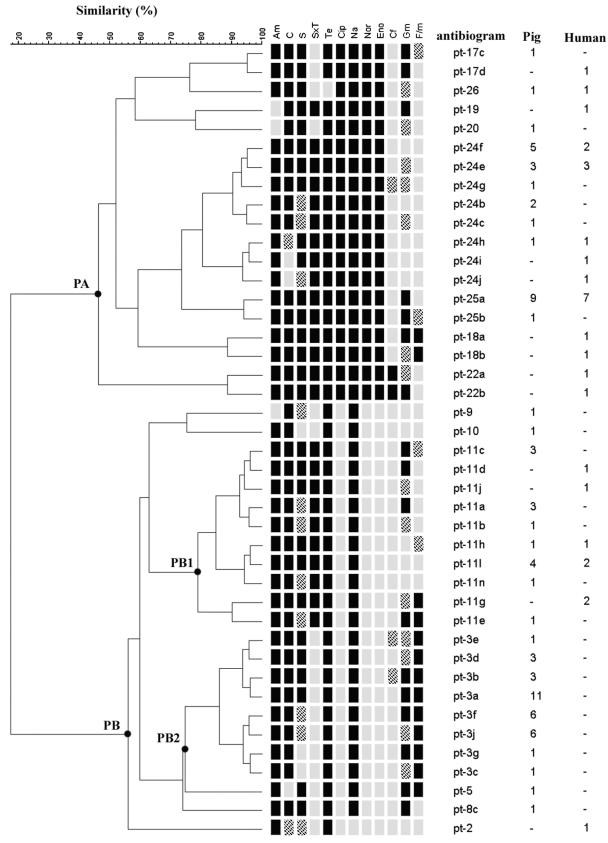
Even though the gt-1a genotype isolates had identical PFGE genotypes, they exhibited quite diverse profiles of susceptibility to the 12 antimicrobial agents tested. A total of 44 susceptibility profiles were identified among the 106 gt-1a isolates. The dendrogram for the susceptibility profiles consisted of two distinct groups, clusters PA and PB (Fig. 2). Clusters PA and PB were separated mainly on the basis of their susceptibilities to the three fluoroquinolones tested in our study. That is, all of the isolates in cluster PA were resistant to the three fluoroquinolones, while those in cluster PB were susceptible to the three drugs. Cluster PB had two major subclusters, subcluster PB1 and subcluster PB2. The response to trimethoprim-sulfamethoxazole was the main difference between the two subclusters. Isolates of cluster PB1 were resistant to trimethoprim-sulfamethoxazole, but those of cluster PB2 were susceptible to the drug. When the relationship between the susceptibility profiles and host origin was considered, 73% (22 of 30) of human isolates of gt-1a were located in cluster PA (fluoroquinolone-resistant cluster), whereas only 34% (26 of 76) of swine isolates of gt-1a were in this cluster. On the basis of such a susceptibility profile analysis with isolates of the same genotype, it was easily observed that the development of drug resistance traits in the genetic strain was more likely to be associated with the host origin. Moreover, the geographic origin of the isolates was also an important factor associated with the susceptibility profile (data not shown). Besides, as shown in Fig. 2, the gt-1a isolates resistant to nalidixic acid were not necessarily resistant to the three fluoroquinolones. However, isolates resistant to any of the three fluoroquinolones were resistant to nalidixic acid and the other two fluoroquinolones.
FIG. 2.
Dendrogram and profiles of susceptibility to 12 antimicrobials for the 106 gt-1a S. enterica serovar Choleraesuis isolates and the numbers of isolates of pig and human origin. Am, ampicillin; C, chloramphenicol; S, streptomycin; SxT, trimethoprim-sulfamethoxazole; Te, tetracycline; Cip, ciprofloxacin; Na, nalidixic acid; Nor, norfloxacin; Eno, enrofloxacin; Cf, cephalothin; Gm, gentamicin; F/m, nitrofurantoin; black box, resistant; grid box, intermediate; gray box, susceptible.
As shown in Table 2, no resistance to ciprofloxacin, enrofloxacin, or norfloxacin was found among the isolates tested before 2000. However, resistance to the fluoroquinolones emerged abruptly in 2000. Since then, high rates of resistance were found among the isolates from both humans and pigs. Several genotypes were identified among the fluoroquinolone-resistant isolates. Among them, gt-1a was still the most prevalent genotype. Among the fluoroquinolone-resistant isolates, 81% (22 of 27) from humans and 87% (26 of 30) from pigs belonged to gt-1a. Since the fluoroquinolone-resistant isolates had closely related PFGE patterns (Fig. 1), they could have evolved from a common ancestor, possibly the gt-1a clone. According to the time of collection of the isolates, the gt-1a strain was present in human and swine populations no later than 1997. Until 1999, human and swine isolates of gt-1a were susceptible to the three fluoroquinolones tested. However, in 2000, most of the gt-1a isolates of both host origins were resistant to the fluoroquinolones, and the rates of resistance remained high from 2000 to 2002. If we accounted only for the isolates recovered from 2000 to 2002, more than 85% of the isolates from humans were resistant, whereas 48% of the isolates from swine were resistant.
TABLE 2.
Genotype distribution of fluoroquinolone-resistant S. enterica serovar Choleraesuis by year from 1997 to 2002
| Source and PFGE subtype | No. of resistant isolates
|
||||||
|---|---|---|---|---|---|---|---|
| 1997 | 1998 | 1999 | 2000 | 2001 | 2002 | Total | |
| Human | |||||||
| gt-2 | 1 | 1 | |||||
| gt-1a | 7 | 8 | 7 | 22 | |||
| gt-1f | 2 | 2 | |||||
| gt-1i | 1 | 1 | |||||
| gt-1k | 1 | 1 | |||||
| Totala | 0/5 | 0/0 | 0/2 | 8/9 | 10/14 | 9/12 | 27/42 |
| Swine | |||||||
| gt-7 | 1 | 1 | |||||
| gt-1a | 8 | 5 | 13 | 26 | |||
| gt-1f | 1 | 1 | |||||
| gt-1h | 1 | 1 | |||||
| gt-1j | 1 | 1 | |||||
| Totala | 0/13 | 0/24 | 0/27 | 8/43 | 6/42 | 16/38 | 30/157 |
The data represent the number of resistant isolates/total number of isolates.
DISCUSSION
The emergence of fluoroquinolone-resistant Salmonella serovar Choleraesuis isolates has become a growing public health concern in Taiwan in recent years. It is mainly a result of the high prevalence of the serovar found among invasive nontyphoid Salmonella serotypes, especially given the fact that only a few alternative therapeutic agents can be used to cure the infections. As reported by Chiu et al. (11), ciprofloxacin resistance was first detected in Salmonella serovar Choleraesuis in 2000 among isolates collected in a medical center located in northern Taiwan. During 2000 and 2001, the fact that more than 40% of the Salmonella serovar Choleraesuis isolates were resistant to ciprofloxacin suggested that an outbreak caused by a resistant strain could have occurred at the time. That speculation is also supported in this study by the results of PFGE genotyping and antimicrobial susceptibility testing of the Salmonella serovar Choleraesuis isolates recovered from humans and swine in different geographic locations from 1997 to 2002. By PFGE genotyping, we identified a genotypic strain, gt-1a, which had been an epidemic strain for humans and pigs at least since 1997. It is speculated that the gt-1a strain could have caused an epidemic before 1997 and that it could be more invasive for humans and swine than other genotypic strains. Consequently, this genotypic strain continued to circulate among its hosts and then developed resistance to fluoroquinolones in response to antimicrobial treatment abuse. Our findings suggest that the fluoroquinolone-resistant Salmonella serovar Choleraesuis strains evolved from a gt-1a clone that emerged in 2000 and that since then has caused widespread infections among humans and pigs.
During the recent epidemic of ciprofloxacin-resistant Salmonella serovar Choleraesuis in humans in Taiwan, pigs were considered the major source of the infection. The major reasons are that they are the primary host for this organism and that enrofloxacin is commonly used as a growth promoter in pig feed and can boost the development of resistance to fluoroquinolones (11, 17, 23). Moreover, the existence of a common genotypic ciprofloxacin-resistant strain from humans and swine is usually considered evidence for the hypothesis. Nowadays, however, molecular techniques are not able to show the direction of transmission, i.e., from humans to pigs or from pigs to humans. Therefore, it is still arguable whether pigs were the source of human fluoroquinolone-resistant Salmonella serovar Choleraesuis strains. In our study, in addition to molecular comparisons of the isolates, further analyses by use of traditional epidemiologic indicators were applied to demonstrate the likelihood of transmission direction. Our data showed that the rates of resistance to ciprofloxacin as well as several other antimicrobials among Salmonella serovar Choleraesuis isolates from humans was much higher than those among isolates from pigs (Table 1). The diverse profiles of susceptibility to the 12 antimicrobials tested found among the 106 identical genotypic (gt-1a) isolates revealed the complexity of the development of resistance traits (Fig. 2). The various susceptibility profile patterns were identified for isolates from different hosts and geographic locations, and these patterns suggest that different levels of antimicrobial use could be the major factor responsible for the development of resistance of the pathogen. In fact, more kinds of fluoroquinolones have been used for the treatment of salmonella infections in humans than in swine. Therefore, the development of ciprofloxacin resistance is more likely to occur in the Salmonella serovar Choleraesuis isolates from human. In this study, we found that human isolates exhibit higher rates of resistance than swine isolates to most of the antimicrobials tested. Therefore, humans cannot be excluded as the source of ciprofloxacin-resistant Salmonella serovar Choleraesuis.
Salmonella serovar Choleraesuis is adapted to the pig host. The bacteria can be shed in feces and remain viable and infective in feces for months or more than a year. Therefore, contaminated swine fecal matter can serve as a reservoir for Salmonella serovar Choleraesuis (16). This organism also causes invasive infections in humans, especially elderly people with underlying diseases (8, 12). It is believed that humans can acquire Salmonella serovar Choleraesuis from the contaminated meat of food animals. However, human infections with Salmonella serovar Choleraesuis also manifests as diarrhea (12), indicating that this organism can be shed in human feces. We have been able to recover three Salmonella serovar Choleraesuis isolates from fecal specimens from 50 human cases with diarrhea (C.-S. Chiou et al., unpublished data). Since only a small proportion of people infected with Salmonella serovar Choleraesuis develops symptoms, transmission from person to person might not be traceable. Therefore, the possibility that Salmonella serovar Choleraesuis is transmitted from person to person or from person to pig cannot be neglected.
Of the isolates tested, 99% were resistant to nalidixic acid but only 29% were resistant to ciprofloxacin, enrofloxacin, and norfloxacin (Table 1). Quinolone resistance can be mediated by target (DNA gyrase and topoisomerase IV) changes and decreased intracellular accumulation resulting from enhanced active efflux pumps and/or a decrease in cell membrane permeability (3, 26). Although the mechanisms for quinolone resistance are very complicated (5, 15, 25), the most frequently reported mechanism for quinolone resistance is point mutations in the gyrase gene (gyrA and gyrB) and the topoisomerase IV gene (parC and parE), especially in the quinolone resistance-determining regions of these genes (17, 19, 25, 27). Nalidixic acid is the primitive form of quinolone. Changes to either the targets or the proteins that control the accumulation and permeation of the antimicrobials could easily result in high levels of resistance to nalidixic acid (15, 17, 25). Ciprofloxacin, enrofloxacin, and norfloxacin belong to the fluoroquinolones. As the derivatives of quinolone, they confer better antimicrobial activity and have better pharmacokinetic performance than nalidixic acid (21). It has been demonstrated that the development of resistance to these modified quinolones is much more complex than that to nalidixic acid. High levels of fluoroquinolone resistance may require multiple amino acid changes in the gyrase and topoisomerase IV sequences, as well as combinations of changes in the specific components, such as active efflux pumps and porins (19, 26). The mechanism of fluoroquinolone resistance has been thoroughly studied by using Salmonella serovar Choleraesuis strains isolated in Taiwan in recent years. The results revealed that almost all of the ciprofloxacin-resistant isolates from humans and pigs have identical amino acid substitutions in codon 83 and codon 87 of gyrA gene (11, 17, 18). These results are concordant with our finding that the fluoroquinolone-resistant Salmonella serovar Choleraesuis strains evolved from a common clone.
In this study, we found that the human isolates had lower rates of resistance to nitrofurantoin. Nitrofurantoin is an antibiotic used to treat urinary tract infections in humans; the reason why the swine Salmonella serovar Choleraesuis isolates have higher rates of percentage to nitrofurantoin remains to be deciphered. In future studies it will be of major interest to investigate whether this antimicrobial agent was widely used for veterinary clinical use in Taiwan.
In conclusion, our study indicated that the cases of human Salmonella serovar Choleraesuis infection in Taiwan from 1997 to 2002 were most likely caused by isolates from a genetically related group which could have evolved from a common genotypic (gt-1a) strain. Infections with the gt-1a Salmonella serovar Choleraesuis group could have contributed to the high prevalence of the serovar among the invasive salmonella isolates detected in humans in Taiwan since 1997 or even earlier. Fluoroquinolone resistance gradually developed from a gt-1a clone and since 2000 has caused widespread infections among humans and pigs. The diverse susceptibility profiles found for the 106 gt-1a Salmonella serovar Choleraesuis isolates demonstrated the complexity of the mechanisms for the development of resistance to antimicrobials. On the basis of our findings of the higher rates of antimicrobial resistance among human isolates than among pig isolates, it is still under debate whether fluoroquinolone-resistant Salmonella serovar Choleraesuis was transmitted from pigs to humans.
Acknowledgments
This research was funded by research grants (grants DOH-92-DC-2029 and DOH-93-DC-2004) from the Department of Health, Executive Yuan, and from Bureau of Animal and Plant Health Inspection and Quarantine, Council of Agriculture, Executive Yuan.
REFERENCES
- 1.Bangtrakulnonth, A., S. Pornreongwong, C. Pulsrikarn, P. Sawanpanyalert, R. S. Hendriksen, D. M. Lo Fo Wong, and F. M. Aarestrup. 2004. Salmonella serovars from humans and other sources in Thailand, 1993-2002. Emerg. Infect. Dis. 10:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrell, R. A. 1987. Isolations of salmonellas from humans and foods in the Manchester area: 1981-1985. Epidemiol. Infect. 98:277-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blondeau, J. M. 2004. Fluoroquinolones: mechanism of action, classification, and development of resistance. Surv. Ophthalmol. 49(Suppl. 2):S73-S78. [DOI] [PubMed] [Google Scholar]
- 4.Brenner, F., and A. C. McWhorten-Murlin. 1998. Identification and serotyping of Salmonella. National Salmonella Reference Laboratory, Centers for Disease Control and Prevention, Atlanta, Ga.
- 5.Cebrian, L., E. Sirvent, J. C. Rodriguez Diaz, M. Ruiz, and G. Royo. 2003. Characterisation of Salmonella spp. mutants produced by exposure to various fluoroquinolones. Int. J. Antimicrob. Agents 22:134-139. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2003. Salmonella surveillance summary, 2002. Centers for Disease Control and Prevention, Atlanta, Ga.
- 7.Chang, C. F., L. C. Chang, Y. F. Chang, M. Chen, and T. S. Chiang. 2002. Antimicrobial susceptibility of Actinobacillus pleuropneumoniae, Escherichia coli, and Salmonella choleraesuis recovered from Taiwanese swine. J. Vet. Diagn. Investig. 14:153-157. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Y. H., T. P. Chen, P. L. Lu, Y. C. Su, K. P. Hwang, J. J. Tsai, H. H. Cheng, and C. F. Peng. 1999. Salmonella choleraesuis bacteremia in southern Taiwan. Kaohsiung J. Med. Sci. 15:202-208. [PubMed] [Google Scholar]
- 9.Chen, Y. H., T. P. Chen, J. J. Tsai, K. P. Hwang, P. L. Lu, H. H. Cheng, and C. F. Peng. 1999. Epidemiological study of human salmonellosis during 1991-1996 in southern Taiwan. Kaohsiung J. Med. Sci. 15:127-136. [PubMed] [Google Scholar]
- 10.Chiu, C. H., T. Y. Lin, and J. Y. Ou. 1999. Predictors for extraintestinal infection of non-typhoidal Salmonella in patients without AIDS. Int. J. Clin. Pract. 53:161-164. [PubMed] [Google Scholar]
- 11.Chiu, C. H., T. L. Wu, L. H. Su, C. Chu, J. H. Chia, A. J. Kuo, M. S. Chien, and T. Y. Lin. 2002. The emergence in Taiwan of fluoroquinolone resistance in Salmonella enterica serotype Choleraesuis. N. Engl. J. Med. 346:413-419. [DOI] [PubMed] [Google Scholar]
- 12.Chiu, S., C. H. Chiu, and T. Y. Lin. 2004. Salmonella enterica serotype Choleraesuis infection in a medical center in northern Taiwan. J. Microbiol. Immunol. Infect. 37:99-102. [PubMed] [Google Scholar]
- 13.Ferris, K., and D. A. Miller. 1990. Salmonella serotypes from animals and related sources reported during July 1989-June 1990, p. 492-504. In Proceedings of the 94th Annual Meeting of the U.S. Animal Health Association. U.S. Animal Health Association, Richmond, Va.
- 14.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35:2977-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giraud, E., A. Brisabois, J. L. Martel, and E. Chaslus-Dancla. 1999. Comparative studies of mutations in animal isolates and experimental in vitro- and in vivo-selected mutants of Salmonella spp. suggest a counterselection of highly fluoroquinolone-resistant strains in the field. Antimicrob. Agents Chemother. 43:2131-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray, J. T., P. J. Fedorka-Cray, T. J. Stabel, and T. T. Kramer. 1996. Natural transmission of Salmonella choleraesuis in swine. Appl. Environ. Microbiol. 62:141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsueh, P. R., L. J. Teng, S. P. Tseng, C. F. Chang, J. H. Wan, J. J. Yan, C. M. Lee, Y. C. Chuang, W. K. Huang, D. Yang, J. M., Shyr, K. W. Yu, L. S. Wang, J. J. Lu, W. C. Ko, J. J. Wu, F. Y. Chang, Y. C. Yang, Y. J. Lau, Y. C. Liu, C. Y. Liu, S. W. Ho, and K. T. Luh. 2004. Ciprofloxacin-resistant Salmonella enterica Typhimurium and Choleraesuis from pigs to humans, Taiwan. Emerg. Infect. Dis. 10:60-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, T. M., Y. F. Chang, and C. F. Chang. 2004. Detection of mutations in the gyrA gene and class I integron from quinolone-resistant Salmonella enterica serovar Choleraesuis isolates in Taiwan. Vet. Microbiol. 100:247-254. [DOI] [PubMed] [Google Scholar]
- 19.Kanematsu, E., T. Deguchi, M. Yasuda, T. Kawamura, Y. Nishino, and Y. Kawada. 1998. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of DNA topoisomerase IV associated with quinolone resistance in Enterococcus faecalis. Antimicrob. Agents Chemother. 42:433-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khakhria, R., D. Woodward, W. M. Johnson, and C. Poppe. 1997. Salmonella isolated from humans, animals and other sources in Canada, 1983-92. Epidemiol. Infect. 119:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kidwai, M., P. Misra, and R. Kumar. 1998. The fluorinated quinolones. Curr. Pharm. Des. 4:101-118. [PubMed] [Google Scholar]
- 22.Kingsley, R. A., and A. J. Baumler. 2000. Host adaptation and the emergence of infectious disease: the Salmonella paradigm. Mol. Microbiol. 36:1006-1014. [DOI] [PubMed] [Google Scholar]
- 23.McDonald, L. C., M. T. Chen, T. L. Lauderdale, and M. Ho. 2001. The use of antibiotics critical to human medicine in food-producing animals in Taiwan. J. Microbiol. Immunol. Infect. 34:97-102. [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests, 7th ed., p. 1-18. NCCLS document no. M2-A7. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 25.Piddock, L. J., V. Ricci, I. McLaren, and D. J. Griggs. 1998. Role of mutation in the gyrA and parC genes of nalidixic-acid-resistant salmonella serotypes isolated from animals in the United Kingdom. J. Antimicrob. Chemother. 41:635-641. [DOI] [PubMed] [Google Scholar]
- 26.Tran, J. H., and G. A. Jacoby. 2002. Mechanism of plasmid-mediated quinolone resistance. Proc. Natl. Acad. Sci. USA 99:5638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trees, D. L., A. L. Sandul, V. Peto-Mesola, M. R. Aplasca, H. B. Leng, W. L. Whittington, and J. S. Knapp. 1999. Alterations within the quinolone resistance-determining regions of GyrA and ParC of Neisseria gonorrhoeae isolated in the Far East and the United States. Int. J. Antimicrob. Agents 12:325-332. [DOI] [PubMed] [Google Scholar]