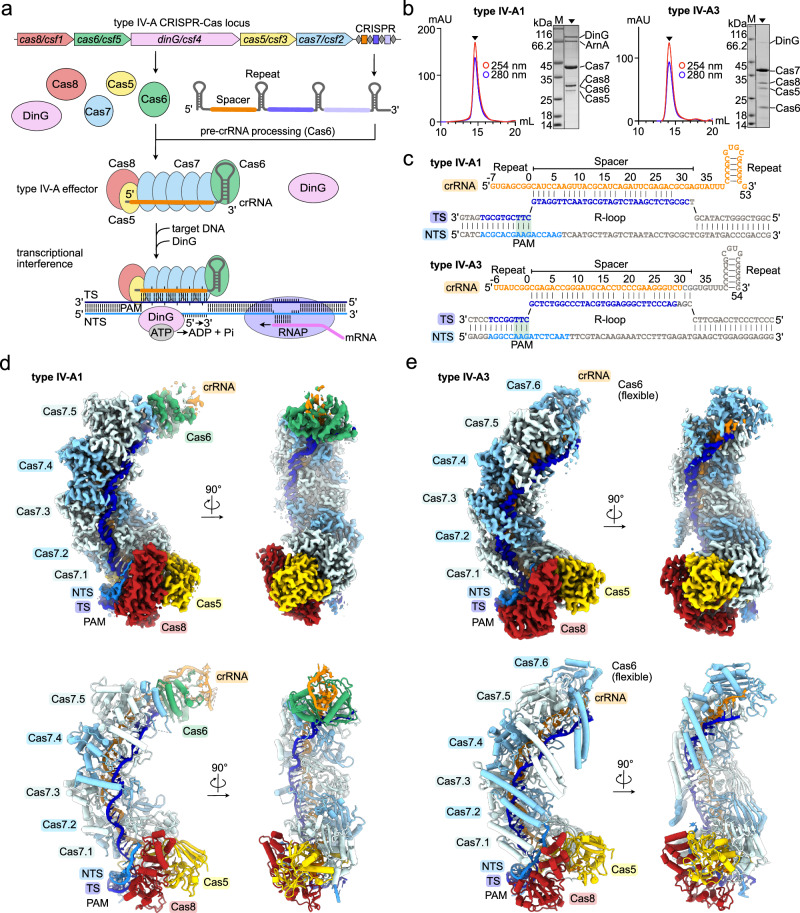
Fig. 1. Structures of types IV-A1 and IV-A3.
a Scheme illustrating a CRISPR-Cas type IV-A locus and its transcriptional interference function. Effector subunits are color-coded according to their identity throughout the manuscript. b Analytical size-exclusion chromatography traces and SDS-PAGE of purified types IV-A1 (left) and IV-A3 (right) effector complexes. ArnA was identified by mass spectrometry as a purification artifact. n = 1. Source data are provided as a Source Data file. c crRNA and DNA substrate sequences of types IV-A1 (top) and IV-A3 (below). Bases resolved in the cryo-EM maps (d, e) are colored orange (crRNA) and blue (DNA). Observed or predicted base pairs are indicated by links between colored or gray bases, respectively. d and e Sharpened experimental cryo-EM maps (top) in two 90°-rotated orientations of the type IV-A1 (d) and type IV-A3 (e) effectors in complex with DNA. Unfiltered experimental and EMReady maps are shown in Supplementary Figs. 2–4. Below: structure models of the type IV-A1 (d) and type IV-A3 (e) effectors in complex with DNA in two 90°-rotated orientations.