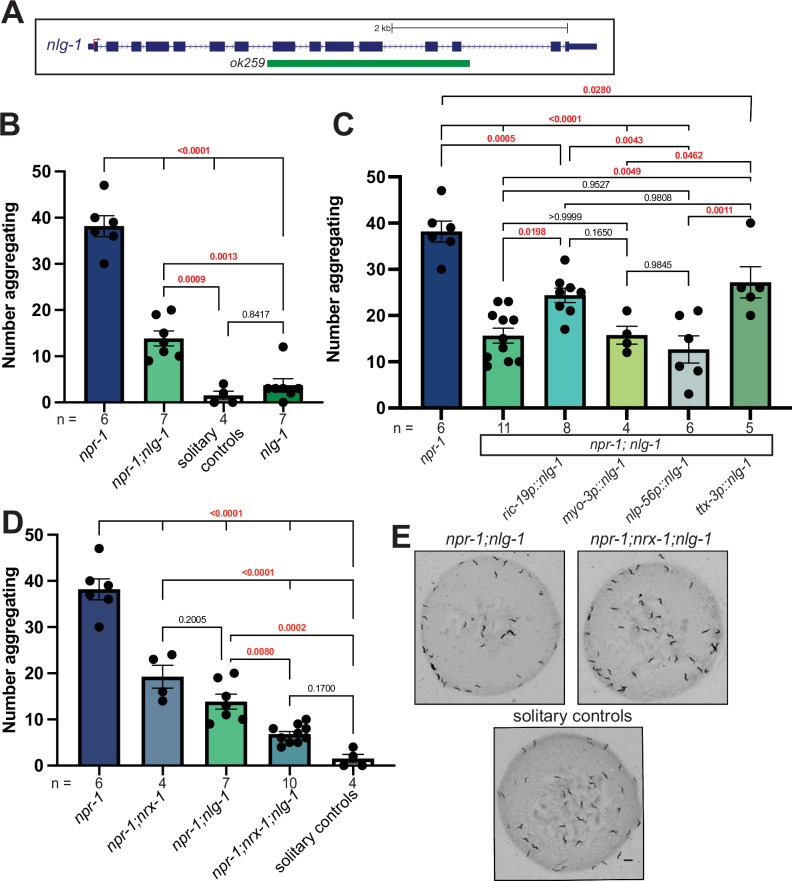
Fig. 3. NLG-1 contributes independent of NRX-1 in aggregation behavior.
A Schematic of C. elegans nlg-1 gene showing the deletion allele assessed. B Graph showing number of aggregating animals in npr-1(ad609), npr-1(ad609);nlg-1(ok259), nlg-1(ok529), and solitary controls. nlg-1 deletion decreased aggregation behavior in npr-1 animals. C Graph showing number of aggregating animals in npr-1(ad609);nlg-1(ok259) mutants with NLG-1 driven by ric-19, myo-3, nlp-56, and ttx-3 promoters and controls. ric-19p expresses in all neurons, myo-3p expresses in muscles, nlp-56p expresses in RMG neurons, and ttx-3p expresses in AIY neurons. D Graph showing number of aggregating animals in npr-1(ad609), npr-1(ad609);nrx-1(wy778), npr-1(ad609);nlg-1(ok259), npr-1(ad609);nrx-1(wy778);nlg-1(ok259), and solitary controls. E Representative images of aggregation behavior in npr-1(ad609);nlg-1(ok259), npr-1(ad609);nrx-1(wy778); nlg-1(ok259) and solitary controls (Scale bar = 1 mm). Data for npr-1 and npr-1;nlg-1 are plotted in 3B, 3C, and 3D. Data for solitary controls are plotted in 3B and 3C. The number of biological replicates (n) are displayed in the figure by order and color, bars show the mean number of aggregating C. elegans, and error bars indicate SEM. One-way ANOVA with Tukey’s post-hoc test was used for comparisons, exact p-values are shown on graphs (red indicates significance, black indicates non-significance). Source data are provided in the Source Data file.