Abstract
Kinetoplastid flagellates are characterized by uniquely massed mitochondrial DNAs (mtDNAs), the kinetoplasts. Kinetoplastids of the trypanosomatid group possess two types of mtDNA molecules: maxicircles bearing protein and mitoribosomal genes and minicircles specifying guide RNAs, which mediate uridine insertion/deletion RNA editing. These circles are interlocked with one another to form dense networks. Whether these peculiar mtDNA features are restricted to kinetoplastids or prevail throughout Euglenozoa (euglenids, diplonemids, and kinetoplastids) is unknown. Here, we describe the mitochondrial genome and the mitochondrial ultrastructure of Diplonema papillatum, a member of the diplonemid flagellates, the sister group of kinetoplastids. Fluorescence and electron microscopy show a single mitochondrion per cell with an ultrastructure atypical for Euglenozoa. In addition, DNA is evenly distributed throughout the organelle rather than compacted. Molecular and electron microscopy studies distinguish numerous 6- and 7-kbp-sized mitochondrial chromosomes of monomeric circular topology and relaxed conformation in vivo. Remarkably, the cox1 gene (and probably other mitochondrial genes) is fragmented, with separate gene pieces encoded on different chromosomes. Generation of the contiguous cox1 mRNA requires trans-splicing, the precise mechanism of which remains to be determined. Taken together, the mitochondrial gene/genome structure of Diplonema is not only different from that of kinetoplastids but unique among eukaryotes as a whole.
Euglenozoa (as defined in reference 13) comprise three morphologically distinct groups, the euglenids, diplonemids, and kinetoplastids. Several types of evidence support the taxonomic cohesion of Euglenozoa. They share discoidal mitochondrial cristae and a characteristic architecture of the flagellar and feeding apparatus (58, 69). Moreover, Euglenozoa add a spliced-leader RNA, a short, capped, noncoding transcript, to all cytoplasmic mRNAs (for reviews, see references 6 and 33). Another feature in common is the modified base “J” (beta-d-glucosyl-hydroxymethyluracil) that occurs in euglenozoan nuclear DNA (16). Most importantly, molecular phylogenies recover the monophyly of Euglenozoa (see, e.g., references 61 and 64).
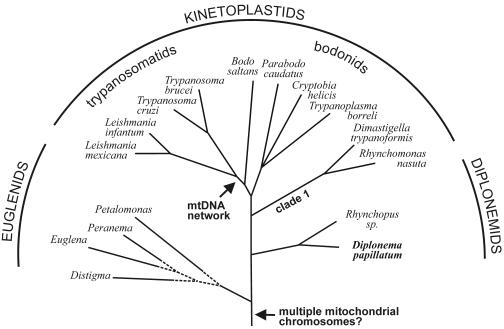
But within Euglenozoa, phylogenetic relationships have been controversial for decades (30, 32, 40, 42, 43, 70). Only recently published molecular phylogenies provide significant support for euglenids diverging most basally within Euglenozoa, followed by the split into two sister groups, the diplonemids and kinetoplastids (59, 60, 61) (Fig. 1). While Euglenozoa as a whole have been viewed historically as one of the most early branching eukaryotic lineages (64), a critical review of statistical support and robustness of global phylogenetic trees questions the basal position of Euglenozoa (50).
FIG. 1.
Schematic phylogenetic tree of Euglenozoa. The topology is taken from studies by Simpson and coworkers (60, 61), where bootstrap support is significant for both the common ancestry of Euglenozoa and the diplonemids and kinetoplastids (but see references 43 and 73). The branching order within euglenids is tentative. Arrows indicate, as proposed in this report, the time points in evolutionary history when multiple mitochondrial chromosomes and mtDNA networks arose in Euglenozoa. Note that Parabodo caudatus was previously designated Bodo caudatus (43).
The largest and best-studied euglenozoan group are kinetoplastids, small colorless flagellates characterized by massed mitochondrial DNAs (mtDNAs) that form stainable structures visible by light microscopy, i.e., the kinetoplasts. Kinetoplastids are subdivided into trypanosomatids and bodonids. Trypanosomatid mtDNAs are composed of interlocked (catenated) networks of “maxicircles” and “minicircles” that are packaged with the aid of proteins into a single, dense disk (74). Maxicircles encode classical mitochondrial genes, most of whose transcripts undergo massive uridine insertion and deletion editing to generate translatable mRNAs. Minicircles encode guide RNAs (gRNAs), the templates for editing pre-mRNAs (reviewed in references 17, 37, and 62). Within bodonids, some taxa have a single kinetoplast similar to trypanosomatids, while others possess multiple mtDNA nodules or DNA distributed diffusely throughout the mitochondrion without visible superstructure. RNA editing activity has been confirmed for two bodonids, Bodo saltans and Trypanoplasma borreli (for a review, see reference 35).
Euglenids are the second largest group within Euglenozoa. The model organism Euglena gracilis has been extensively studied, at the level of both the nuclear and the chloroplast genome, whereas euglenid mtDNA seems to be intractable with present methods. So far, cox1 (coding for cytochrome oxidase subunit 1) is the only mitochondrial gene cloned and sequenced for E. gracilis (68, 76). The structure of the mitochondrial genome as a whole remains unknown.
Diplonemids, the third and smallest group within Euglenozoa, contains a total of two genera, Rhynchopus and Diplonema (30, 55). First described in 1914 (24), Diplonema was rediscovered in the late 1960s (55), designated Isonema, and misplaced within euglenids (52). Most diplonemids inhabit marine ecosystems (63) and, similar to their kinetoplastid sister group, they include pathogenic taxa. Some Rhynchopus species parasitize diatoms (54) and others lobster (73), while Diplonema not only has been reported to infest clams (29) but also has been associated with Cryptocoryne disease, which causes the sudden decay of aquarium plants (65).
The mitochondrial genome of diplonemids is virtually uncharacterized. The only molecular data currently available are a partial cox1 cDNA sequence of Diplonema papillatum (40), but the subcellular location of the corresponding transcript or its gene is not known. We initially attempted to characterize Diplonema mtDNA using whole genome shotgun sequencing within the context of the Organelle Genome Megasequencing Program (http://megasun.bch.umontreal.ca/ogmpproj.html). But sequence assembly of the presumptive mtDNA was highly ambiguous, due to extensive sequence repeats, and not a single known mitochondrial gene was discernible in the ∼20 kbp that was sequenced (G. Burger, B. F. Lang, and M. W. Gray, unpublished results). This report describes the approach that allowed us to identify and characterize mtDNA for Diplonema, thus laying the groundwork for the development of new strategies to sequence its complete mitochondrial genome and for the discovery of an unprecedented mitochondrial gene and genome structure for this organism.
MATERIALS AND METHODS
Strains and protist culture.
D. papillatum (ATCC 50162) was obtained from the American Type Culture Collection. The organism was cultivated axenically at ∼22°C in artificial seawater and enriched with 1% fetal bovine serum (Wisent) and 0.1% tryptone.
Light microscopy.
Cells were resuspended in phosphate-buffered saline (PBS) at a concentration of 107 cells/ml, and the suspension was spotted onto poly-l-lysine-coated slides. After the cells were allowed to adhere for 30 min in a humidity chamber, the slides were submerged for 5 min in methanol. Fixation was stopped by washing the slides with PBS, and cells were stained with 0.1 μg/ml DAPI (4′,6′-diamidino-2-phenylindole; a DNA-specific fluorescence dye) in PBS for 3 min. For staining with the fluorescence dye YOYO1 (oxazole yellow dimer; Molecular Probes), cells were attached to slides as described above, fixed in PBS with 4% paraformaldehyde for 3 min, washed twice in PBS, and permeabilized in PBS with 0.5% Tween 20 for 30 min. After a single wash in PBS containing 0.05% Tween 20 for 5 min, cells were incubated with 10 nM YOYO1 for 20 min and rinsed twice with PBS. The DAPI- and YOYO1-stained cells were mounted in antifade reagent [0.233 g 1,4-diazabicyclo-(2,2,2) octane; 1 ml 0.2 M Tris-HCl, pH 8.0; and 9 ml glycerol], and the slides were examined with a Zeiss Axioplan 100 microscope.
Electron microscopy.
For transmission electron microscopy, cells were washed twice in PBS and fixed in the same buffer with 2.5% glutaraldehyde for 1 h at 4°C and then washed in PBS, fixed with 2% osmium tetroxide in the same buffer, and finally washed in distilled water. After dehydration in graded series of ethanol, the cells were embedded in Epon-Araldite. Thin sections of embedded cells were stained with lead citrate and uranyl acetate and examined under a JEOL JEM 1010 microscope. The images of adjacent sections (60 to 70 nm thick) were registered interactively by translations and rotations. Contours of structures to be reconstructed were outlined manually in distinct colors. Volumetric models of the structures were generated by plotting the contours in successive planes of the three-dimensional (3D) data sets. To remove irregularities due to deformation of microscopic sections, the data sets were resampled and smoothed. Surface models of the structures were constructed as triangulated isosurfaces of the data by the marching cubes algorithm (28). The final model was generated by combining the colored surfaces. Model images were obtained by rendering the resulting 3D scenes using the OpenGl library.
For electron microscopy studies of the topology and size of DNA molecules, Diplonema mtDNA was prepared by the cytochrome c method and spread and recorded as described earlier (36). The contour lengths of the circular chromosomes were measured from prints using a HIPAD digitizing tablet (Houston Instruments). The sizes determined were 2.05 ± 0.05 μm and 2.35 ± 0.05 μm. The precise magnification was determined by replica granting (Balzers). Plasmid pBR322 served as a size standard for the conversion from μm to kbp.
Isolation of mitochondria and mtDNA extraction.
Cells of D. papillatum were resuspended in STE buffer (0.6 M sorbitol, 50 mM Tris-HCl, and 5 mM EDTA [pH 7.4]) and broken mechanically by shaking with glass beads. The mitochondria were collected after three differential centrifugation steps using a Sorvall SS34 rotor, as follows: two times at 8,000 rpm (7,650 × g) for 10 min and once at 18,000 rpm (38,700 × g) for 20 min. We confirmed that this subcellular fraction is highly enriched in mitochondria by electron microscopy, which revealed the typical double-membrane-bounded vesicles (data not shown). The mitochondrion-containing pellet was resuspended in STE buffer and subsequently lysed in the presence of 1% (vol/vol) sodium dodecyl sulfate (SDS) and 100 μg/ml proteinase K. The SDS was eliminated by NaCl precipitation, the lysate was phenol-chloroform extracted, and the DNA was precipitated by ethanol. Nuclear DNA was isolated from the pellet obtained after the first centrifugation step. For a more thorough purification of mitochondria, the mitochondrion-containing pellet was resuspended in 15% sucrose (and 0.1 mM EDTA) and layered on top of a sucrose step gradient (25, 35, 45, and 65% sucrose). Centrifugation was carried out by using a swing-out rotor (Sorvall TH641) at 38,000 rpm (224,000 × g) for 90 min. The mitochondrial band, which formed between the layers of 35% and 45% sucrose, was collected and used directly for mtDNA extraction. The overall mtDNA patterns (monitored by agarose gel electrophoresis) before and after the gradient were identical.
Pulsed-field gel electrophoresis.
Whole-chromosome sized DNAs were prepared by using agarose plugs, at a concentration of 107 cells/ml. Electrophoresis was carried out in a contour-clamped homogeneous electric field electrophoresis apparatus (CHEF-DR III; Bio-Rad) in 1% agarose (SeaKem) and 1× Tris-acetate-EDTA buffer at 16°C for 11 h, using a linear switch time ramp (1 to 6 s) at an angle of 120° and a voltage of 6 V/cm. After migration, the gel was stained with ethidium bromide.
Isolation and fractionation of total DNA.
Cells from a fresh culture were washed in STE buffer and lysed in the presence of 1% SDS and 100 μg/ml proteinase K for 1 h at 50°C. The SDS was eliminated by NaCl precipitation. Total DNA was purified according to two different methods; either (i) the lysate was phenol-chloroform extracted and, subsequently, the DNA was ethanol precipitated, or (ii) DNA was ethanol precipitated and extracted by spooling around a Pasteur pipette. The second method allows extraction without the centrifugation that might break large DNA molecules. Total DNA was fractionated according to A+T content using CsCl-Hoechst 33258 equilibrium density centrifugation in an NVTi65 rotor at 55,000 rpm for 16 h.
Southern hybridization.
Electrophoresis and blotting were performed by standard procedures. The DNA probes were labeled by random priming in the presence of [α-32P]dATP (Perkin Elmer). Hybridization was performed at 65°C in 1.5× SSPE (3.6 M NaCl, 0.2 M sodium phosphate, and 20 mM EDTA [pH 7.4]) plus 1% SDS, 0.5% dry milk, and 0.1 mg/ml salmon sperm.
PCR amplification.
N-terminal and C-terminal fragments of cox1 were amplified from total or mtDNA using primer set 1 and 2 and primer set 7 and 8, respectively. Different combinations of primers 3 to 8 were used to amplify internal fragments. Primer sequences (5′ to 3′) are as follows: primer 1, GCACTGCTAGTAGGTATCAT; primer 2, CTCTGGGTGCCCGAAGACC; primer 3, CGCTCTACAACATGCTGGGC; primer 4, CCAGGAGAACACCTTGATGGA; primer 5, CCTCGTAATGCATGACAGCT; primer 6, GGTATCCATCAGGTGCATCT; primer 7, CCTGTGGTATACTGGTAGTC; and primer 8, CCTCCAGATGCATGGATGCT. Assuming a gene that is colinear with the cDNA, the expected lengths of the PCR products are 150 bp for primers 1 and 2, 1,006 bp for primers 1 and 8, 171 bp for primers 3 and 4, 543 bp for primers 3 and 6, 771 bp for primers 3 and 8, 500 bp for primers 5 and 8, and 228 bp for primers 7 and 8.
RT-PCR.
Total RNA was extracted using TRIzol. First-strand cDNA synthesis by AMV reverse transcriptase (Boehringer) was primed by an anchored dT oligonucleotide, followed by PCR-amplification using cox1 primers listed above. Alternatively, first strand synthesis was primed by the 3′-distal primer 8, and amplification proceeded by using primers 1 and 8 or 3 and 8. The expected lengths of reverse transcriptase (RT)-PCR products are as listed above.
DNase I digestion.
One μg of mtDNA was digested with 1 × 10−3, 2 × 10−3, or 4 × 10−3 units of DNase I (Fermentas) for 6 min at 4°C and then stopped by the addition of EDTA (50 mM).
DNA sequencing and sequence analysis.
PCR products were purified by spin columns (Millipore). DNA sequencing was performed with an MJ automated sequencer, using the cycle-sequencing/dye termination chemistry (BigDye Terminator 3.1; ABI). Sequence similarity searches were conducted locally (FASTA) (48) against data retrieved from the Organelle Genome Database at http://megasun.bch.umontreal.ca/gobase/ and by remote BLAST searches at the National Center for Biotechnology Information (1).
Nucleotide sequence accession number.
The DNA sequence reported in this paper was deposited in GenBank under accession no. AY686226.
RESULTS
Cellular ultrastructure and mtDNA packaging in Diplonema.
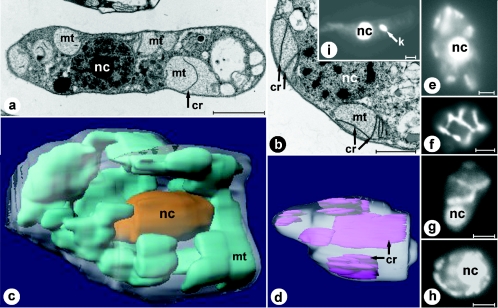
Transmission electron micrographs of both longitudinally and cross-sectioned D. papillatum cells show several large mitochondrial structures (Fig. 2a and b), which are, in fact, branches of a single organelle, as demonstrated later. The mitochondrial lumen is homogenously electron transparent and contains few, exceptionally large and irregularly arranged cristae, which are flat, uniformly thick (∼25 nm), unbranched, and only rarely attached to the mitochondrial inner membrane. Although several mitochondria have been cross-sectioned completely, we did not detect kinetoplast-like bodies (35), as seen with the control (Trypanosoma brucei) (Fig. 2i).
FIG. 2.
Ultrastructure of D. papillatum mitochondria. Electron microscopy (a and b), light microscopy (e to i), and 3D reconstruction (c and d) based on electron microscopy images. Electron micrographs of longitudinally sectioned (a) and cross-sectioned (b) Diplonema cells. Prominent flat cristae are visible in the organellar lumen. (c) 3D reconstruction of the whole cell, based on 48 consecutive sections. The single mitochondrion is shown in cyan, and the posteriorly located oval nucleus is shown in gold. (d) 3D reconstruction of a large compartment of the mitochondrion, based on 25 consecutive sections. Cristae are shown in purple. (e to g) YOYO1-stained D. papillatum cells focused on the nucleus (e and g) and the cell periphery (f). (h) DAPI-stained D. papillatum cell. (i) Control staining (YOYO1) of the kinetoplastid Trypanosoma brucei. Bars, 2 μm (a), 0.5 μm (b), and 2.5 μm (e to i). nc, nucleus; mt, mitochondrion; k, kinetoplast; cr, cristae.
3D reconstruction of serially sectioned Diplonema cells reveals a single, highly branched mitochondrion, anastomosing throughout the cell, but mostly located peripherally (Fig. 2c). Mitochondrial cristae, which protrude irregularly into the lumen, exhibit a variable but overall exceptionally large size (up to 2 μm long and 1 μm wide), lamellar shape, and longitudinal rather than transversal arrangement (Fig. 2d). Both nucleic acid-staining dyes used, YOYO1 (Fig. 2e to g) and DAPI (Fig. 2 h), light up large amounts of mtDNA meandering through the cell. To precisely colocalize DNA and organelle, we attempted to label the mitochondrion with MitoTracker in whole cells. But this experiment failed, possibly due to insufficient membrane permeability. Nevertheless, the pattern of DNA distribution within the cell, as monitored by fluorescence microscopy, is fully consistent with the spatial mitochondrial morphology obtained from 3D reconstruction. This provides clear evidence that mtDNA is uniformly distributed in the mitochondrial branchwork and not compacted in a single kinetoplast, as in trypanosomatids.
Purification and identification of mtDNA.
Employing degenerate primers for RT-PCR, Maslov and colleagues isolated a partial cox1 cDNA of D. papillatum, which includes ∼1/2 of the expected coding sequence starting at a position corresponding to residue 246 of the T. brucei protein (40). This cDNA probe hybridized to a G+C-rich DNA fraction of Diplonema, which the authors considered to be mtDNA, because cox1 is mitochondrion encoded in all species examined to date. However, several considerations suggest the use of caution about such an extrapolation. One is a recent report about the unexpected cytoplasmic localization of typical chloroplast genes in dinoflagellates (31). Moreover, high G+C content is unusual for mtDNA but rather typical for nuclear DNA. Finally, the cox1 cDNA was obtained from a polyadenylated mRNA fraction, while mitochondrial mRNAs of most eukaryotes, except mammals, apicomplexans, and kinetoplastids, do not have A-tails.
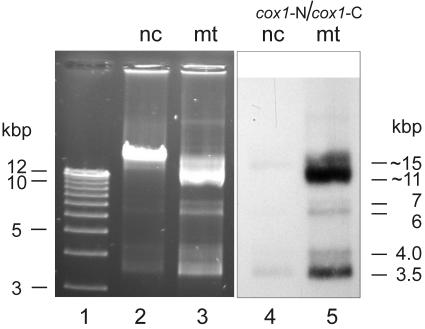
To identify the mtDNA of Diplonema beyond doubt, we gently disrupted the cells mechanically, separated mitochondria from nuclei and other subcellular structures by kinetic step-gradient centrifugation, and extracted the DNA from each subcellular fraction (see Materials and Methods). In parallel, total cellular DNA was separated by CsCl-bisbenzimide equilibrium centrifugation, which yielded two discrete bands, one more A+T rich, the other more G+C rich. Gel electrophoresis shows that the A+T-rich material corresponds to the DNA recovered from nuclei (Fig. 3, lane 2), whereas the G+C-rich material corresponds to mtDNA (Fig. 3, lane 3). In Southern hybridization, labeled PCR-generated fragments of Diplonema cox1 cDNA indeed hybridize to the DNA extracted from mitochondria (Fig. 3, lane 5). Moreover, digestion with restriction enzymes demonstrates that the G+C-rich DNA species is much less complex that the A+T-rich one (data not shown). Together, these results confirm that in Diplonema, the more G+C-rich DNA fraction represents the mitochondrial genome.
FIG. 3.
Identification of D. papillatum mtDNA. Agarose gel electrophoresis of DNA from different subcellular fractions. Electrophoresis was performed in the presence of ethidium bromide. Lane 1, size markers (1 kb plus ladder); lane 2, DNA isolated from the nuclear subcellular fraction (nc). The pattern is identical with that of the A+T-rich band from CsCl-bisbenzimide equilibrium centrifugation (data not shown). Lane 3, DNA extracted from the mitochondrial subcellular fraction (mt). The pattern is identical with that of the G+C-rich band from CsCl-bisbenzimide equilibrium centrifugation (data not shown). Lanes 4 and 5, Southern hybridization of blotted lanes 2 and 3, using as a probe a mixture of cox1 N-terminal and C-terminal fragments (cox1-N and cox1-C; see Fig. 7A) generated by PCR. Note that the nuclear fraction is slightly cross-contaminated by mtDNA, as revealed by staining (lane 2) and hybridization (lane 4). Moreover, a minute proportion (<1/1,000) of a DNA species migrating at ∼23 kbp is seen in all preparations and most likely corresponds to covalently closed tandem homodimers of mitochondrial chromosomes, molecules well known from other systems with circular mtDNA, such as mammals (14) and Bodo species (4, 25). Sizes indicated to the right of lane 5 correspond to the apparent sizes of chromosomes, which are different from their true sizes when molecules are not linear (see text).
Shape and conformation of mtDNA.
From their analyses of restriction enzyme digests, Maslov and colleagues concluded that D. papillatum mtDNA “is probably composed of several types of circles, [whose] exact size, number of types and topological arrangements remain to be investigated” (40). We determined the physical properties of this genome by the following experiments.
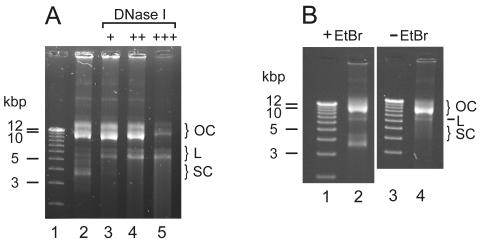
Electrophoresis of purified Diplonema mtDNA on agarose gels (in the presence of ethidium bromide) yielded a complex pattern of three pairs of bands, with apparent sizes of 3.5 and 4 kbp, 6 and 7 kbp, and 11 and 15 kbp, present in unequal stoichiometry (see Fig. 3, lane 3). To test whether these multiple bands are due to two circular molecules occurring in three different conformations or, rather, to six molecules of different sizes, we incubated mtDNA for various periods with the nicking enzyme DNase I. In the case of circular, supercoiled DNA, an increasing number of nicks would first abolish the supercoiled conformation and then linearize the circle, followed by fragmentation and final degradation of this linear form (15). Figure 4A illustrates that treatment of Diplonema mtDNA with DNase I eliminates progressively the bands at 3.5 and 4 kbp followed by those at 11 and 15 kbp, proceeding gradually by first enhancing and then diminishing the bands at 6 and 7 kbp. This behavior is consistent with two molecule classes, of 6 kbp and 7 kbp, present in the following three different conformations: (i) linear (migrating at 6 kbp and 7 kbp according to their size), (ii) relaxed circular (migrating at 11 and 15 kbp), and (iii) supercoiled, covalently closed-circular (migrating at 3.5 and 4.0 kbp). In the present paper, the two size classes of molecules are referred to as class A (6-kbp) and class B (7-kbp) chromosomes.
FIG. 4.
Conformation of D. papillatum mitochondrial chromosomes. OC, relaxed closed circles; L, linear forms; SC, supercoiled, covalently closed circles. (A) incubation of mtDNA in the presence of DNase I. Lane 1, size markers as in Fig. 3; lane 2, control without DNase I treatment; lane 3 to 5, incubation with 1 × 10−3 units (+), 2 × 10−3 units (++), and 4 × 10−3 units (+++) of DNase I for 6 min. (B) Electrophoretic migration in the presence (lane 2) and absence (lane 4) of ethidium bromide. Lanes 1 and 3, size markers as in Fig. 3.
Ethidium bromide induces the supercoiled conformation in relaxed closed DNA circles by intercalation between two successive base pairs. This unwinds the DNA double helix and produces negative supercoils (51). To test whether supercoiling is secondarily induced by staining, we performed electrophoretic separation in the absence of this dye. Indeed, the fast-migrating bands are absent (Fig. 4B, lane 4), demonstrating that in vivo, the major portion of Diplonema mtDNA is in monomeric, relaxed circular conformation.
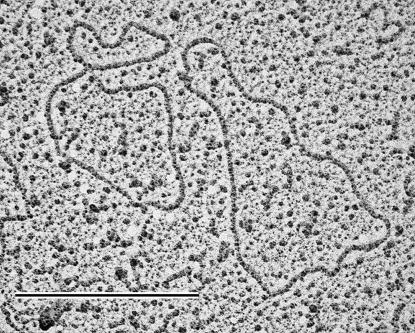
Electron microscopy captures visually the size and topology of Diplonema mitochondrial chromosomes (Fig. 5). For this experiment, DNA was isolated in the absence of intercalating agents. Among 61 screened DNA molecules, >95% are circular and >85% are in clearly relaxed conformation; 54% measured 6.0 to 6.3 kbp (class A) and 46% measured 7.0 to 7.3 kbp (class B). These results are in full agreement with the molecular data described above.
FIG. 5.
Electron microscopy analysis of Diplonema mtDNA. Circular chromosomes in relaxed conformation. Left, class A chromosome (contour length corresponding to 6.0 to 6.3 kbp); right, class B chromosome (contour length corresponding to 7.0 to 7.3 kbp). Bar, 0.5 μm.
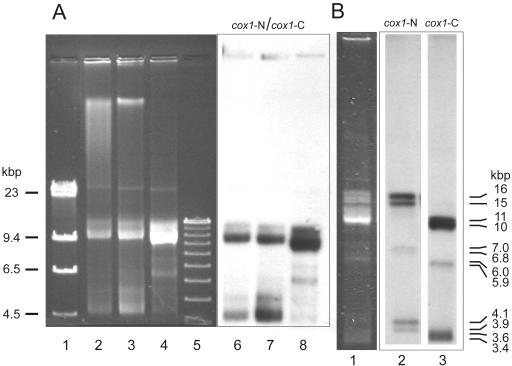
With sizes of only 6 and 7 kbp, Diplonema mitochondrial chromosomes are unusually small. To investigate the potential presence of large mtDNA molecules that may not have entered the gel during conventional electrophoresis, we used pulsed-field gel electrophoresis. Under conditions separating molecules of several hundred kbp, no significant high-molecular band in mtDNA is discernible, either by ethidium bromide-staining or by Southern hybridization (Fig. 6A, lanes 4 and 8). This result shows that Diplonema mtDNA does not include detectable amounts of larger (maxicircle-like) molecules.
FIG. 6.
Structure of the D. papillatum cox1 gene. (A) Pulsed-field separation of DNA. Lane 1, size marker lambda HindIII; lane 2, total DNA purified by spooling; lane 3, total DNA purified by phenol-chloroform extraction; lane 4, mtDNA isolated from the mitochondrial subcellular fraction; lane 5, size marker as in Fig. 3; lanes 6 to 8, blotted lanes 2 to 4, hybridized with a mixture of cox1-N and cox1-C as a probe (see Fig. 7A). (B) Southern hybridization of mtDNA with two different cox1 probes. Lane 1, mtDNA separated by agarose gel electrophoresis in the presence of ethidium bromide; lane 2, hybridization of blotted lane 1 with cox1-N (N-terminal fragment); lane 3, hybridization of blotted lane 1 with cox1-C (C-terminal fragment) (see Fig. 7A). Apparent molecule sizes (see legend to Fig. 3) are indicated to the right of lane 3. For information on the minute band migrating at ∼23 kbp, see legend to Fig. 3. Note that due to the particular migration conditions in this experiment, electrophoretic mobilities of DNA species differ from those shown in Fig. 3 and 4.
Similarity of class A and B chromosomes and localization of cox1.
To test whether the two chromosome classes have sequence in common, we performed Southern hybridizations with three different probes, complete A-class and B-class chromosomes and a 4-kbp genomic cox1 clone of Diplonema, which was isolated and provided to us by D. Maslov. We sequenced this clone and discovered a ∼220-bp-long stretch of significant sequence similarity to cox1, corresponding to amino acids ∼193 to 268 of the T. brucei protein. This stretch of cox1, referred to below as cox1-N, is bounded by highly repetitive sequence (GenBank accession no. AY686226). Each of the above three probes hybridized equally strongly to all Diplonema mtDNA bands (data not shown), showing that the two chromosomes share significant sequence similarity.
The two chromosome classes also include distinct regions, as seen in the following Southern hybridization results. Probes for this experiment contained solely cox1 regions, which were obtained by PCR amplification from mtDNA: cox1-N (see above) and a most C-terminal cox1 portion designated cox1-C (corresponding to residues 477 to 543 of the T. brucei protein; for primers, see Fig. 7A). Surprisingly, the cox1-N probe recognized exclusively class B molecules (Fig. 6B, lane 2), whereas the cox1-C probe specifically bound to class A chromosomes (Fig. 6B, lane 3). In addition, Southern hybridization reproducibly resolved double bands of A and B chromosomes. This implies the presence of two versions each of chromosomes carrying cox1-N and cox1-C sequences, differing in size by 100 to 200 bp.
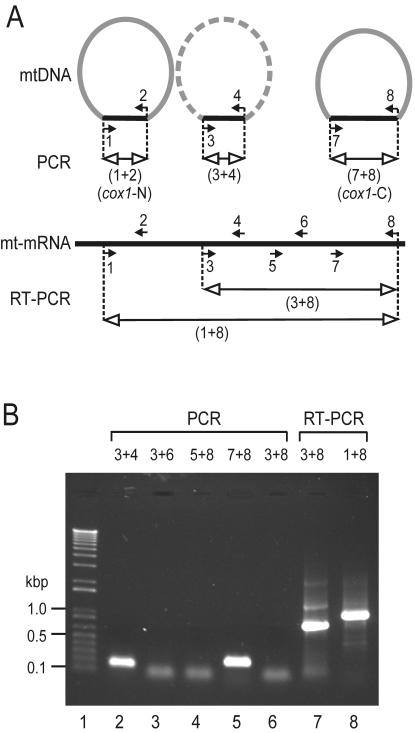
FIG. 7.
PCR amplification of the D. papillatum cox1 gene. (A) Schematic representation of the cox1 sequence and PCR primers. Semicircular forms indicate mtDNA chromosomes, with the cox1 coding region in black and the rest of the chromosome in gray. The existence of the corresponding chromosome remains to be demonstrated (symbolized by the dashed arc), but the cox1 region has been obtained by PCR. The chromosome depicted on the left gave rise to the genomic cox1 clone, which was isolated by D. Maslov and sequenced by us (this report). Filled small arrows with numbers indicate primers and their orientations. Lines with open arrowheads depict PCR products. The portion of mitochondrial mRNA between primers 3 and 8 corresponds to the published cox1 cDNA sequence (GenBank accession no. AF119813 [40]). (B) Electrophoretic separation of PCR products. Lane 1, size marker as in Fig. 3; lanes 2 to 6, mtDNA was PCR amplified using different primer pairs as indicated above the lanes. The numbers refer to the primers in Fig. 7A. Lanes 7 to 8, total RNA was reverse transcribed and PCR amplified using primer set 3 and 8 and primer set 1 and 8, respectively. The diffuse bands migrating at ∼50 bp in lanes 3, 4, 6, and 7 are most likely primer dimers, as they also occur in control experiments without DNA (not shown). For primer sequences, see Materials and Methods.
With the discovery of N-terminal and C-terminal portions of Diplonema cox1 residing on separate mitochondrial chromosomes, the question arises whether a low abundant, continuous cox1 exists as well. Therefore, we conducted PCR experiments with total DNA, using diverse primer pairs to cover different portions of the hypothetical cox1 coding region (Fig. 7A). Figure 7B shows that only amplification of cox1 segments shorter than 300 bp was successful. As a positive control, we performed RT-PCR with total RNA using far distant primers, which yielded products of the predicted sizes (Fig. 7B, lanes 7 and 8) and expected sequence (identical to the cox1 cDNA sequence; GenBank accession no. AF119813 [40]). This demonstrates that instead of a contiguous cox1 gene, Diplonema possesses several ∼250-bp cox1 regions scattered across multiple chromosomes.
DISCUSSION
Atypical ultrastructure of Diplonema mitochondria.
Our 3D reconstructions show for the first time that D. papillatum contains a single mitochondrion per cell, as do kinetoplastids. Apart from that, Diplonema mitochondria appear to be highly atypical for Euglenozoa. Compared to the threadlike mitochondria with numerous tiny and discoidal mitochondrial cristae in kinetoplastids and euglenids (9, 18; see also reference 49), the organelle of D. papillatum is amoeboid in structure and encloses scarce, giant, and flat mitochondrial cristae (see also reference 58). Our study provides new evidence for the notion that Diplonema is at odds with the distinctive discoidal mitochondrial cristae of “Discicristata” (Euglenozoa and Heterolobosea) (47).
Our fluorescence microscopy studies show that, in contrast to the kinetoplast DNA (kDNA) of trypanosomatids, Diplonema mtDNA is distributed diffusely throughout the organelle. A comparable mtDNA distribution occurs sporadically in bodonids (pan-kDNA and mega-kDNA), but the large majority has mtDNA molecules assembled into discrete nodules (poly-kDNA) or a single bundle-like structure (pro-kDNA) (72; see compilation in Table 1).
TABLE 1.
Architecture and gene content of diplonemid and kinetoplastid mtDNAs
| Taxon | mtDNA distributiona | kDNA networkb | Chromosome type | Size (kbp) | Shape/ conformation | Coding content(s) | RNA editing | Source or reference |
|---|---|---|---|---|---|---|---|---|
| Diplonemida | ||||||||
| D. papillatum | Pan-kDNA-like | No | A-class | 6 | Circular | Genes, gRNAs | NDc | This study |
| B-class | 7 | Relaxed | Genes, gRNAs | ND | ||||
| Kinetoplastida- trypanosomatids | ||||||||
| Crithidia fasciculata | Eu-kDNA | Yes | Minicircle | 2.5 | Circular | gRNAs | Yes | 75 |
| Maxicircle | 38 | Relaxed | Genes, gRNAs | |||||
| L. tarentolae | Eu-kDNA | Yes | Minicircle | 0.9 | Circular | gRNAs | Yes | 67 |
| Maxicircle | 21 | Relaxed | Genes, gRNAs | |||||
| Phytomonas serpens | Eu-kDNA | Yes | Minicircle | 1.5 | Circular | gRNAs | Yes | 38 |
| Maxicircle | 27-36 | Relaxed | Genes, gRNAs | |||||
| Trypanosoma avium | Eu-kDNA | Yes | Minicircle | 10 | Circular | ND | ND | 77 |
| Maxicircle | ND | Relaxed | ND | |||||
| T. brucei | Eu-kDNA | Yes | Minicircle | 1.0 | Circular | gRNAs | Yes | 44 |
| Maxicircle | 21-27 | Relaxed | Genes, gRNAs | |||||
| Trypanosoma vivax | Eu-kDNA | Yes | Minicircle | 0.5 | Circular | ND | Yes? | 7 |
| Maxicircle | 23 | Relaxed | ND | |||||
| Kinetoplastida-bodonids | ||||||||
| P. caudatus | Pan-kDNA? | No | Minicircle | 10-12 | Supercoiled | ND | Yes | 25 |
| Maxicircle | 19 | ND | ND | |||||
| B. saltans | Pro-kDNA | No | Minicircle | 1.4 | Circular | gRNAs | Yes | 3 |
| Maxicircle | 70 | Relaxed | Genes, gRNAs | |||||
| Cruzella marina | Poly-kDNA | No | Minicircle | 2.0 | Circular | gRNAs? | ND | 78 |
| Relaxed | ||||||||
| Maxicircle | ND | ND | ND | |||||
| Cryptobia helicis | Pan-kDNA | No | Minicircle | 4.2 | Supercoiled | gRNAs? | ND | 36 |
| Maxicircle | 43 | ND | Genes | |||||
| Dimastigella trypaniformis | Poly-kDNA | No | Minicircle | 1.45 | Circular Relaxed | gRNAs? | ND | 66 |
| Maxicircle | ND | ND | ND | |||||
| Trypanoplasma borreli | Mega-kDNA | No | Minicircle | 200 | Linear 1 kbp Tandem rep. | gRNAs | Yes | 34, 39 |
| Maxicircle | 80 | ND | Genes |
Determined by DNA staining in whole cells.
Determined by electron microscopy and molecular studies of DNA.
ND, not determined.
Unique mitochondrial gene organization in Diplonema.
DNA sequencing, Southern hybridization, and PCR experiments provide evidence that D. papillatum mtDNA contains at least three distinct ∼250 bp-long stretches of cox1 sequence, corresponding approximately to residues 193 to 247, 269 to 326, and 477 to 543 of the Trypanosoma protein. Each of these stretches is located on a separate chromosome; a continuous cox1 coding region was not detected. Two different scenarios could explain this finding. Reminiscent of the situation for kinetoplastids, the observed cox1 stretches might represent gRNA genes involved in editing of heavily encrypted transcripts. However, kinetoplastid gRNA genes are 5 to 10 times shorter than the Diplonema cox1 stretches, and their regions of sequence identity with the mature transcript are only 5 to 15 bp long. Therefore, it is much more likely that the ∼250-bp cox1 stretches are separate coding portions of a discontinuous cox1 gene.
Discontinuous genes are widespread in organelle genomes outside Euglenozoa and also occur in nuclear genomes. Four types of discontinuous genes can be distinguished. Most abundant are intron-containing genes, whose intervening sequences are removed from pre-mRNA by cis-splicing. A second type involves exons that are encoded in distant locations or even on different strands of the chromosome and that are transcribed separately, each with parts of flanking introns. The precise linking of these discrete precursor transcripts to form a mature RNA proceeds by trans-splicing. In organelles, best known from plant mitochondria, this step is mediated by the distinct secondary structure folding of group II introns (41), while trans-splicing in the nucleus, first described for T. brucei, involves spliceosomes (for reviews, see references 6, 33, and 57). Another type of discontinuous genes involves DNA splicing that generates secondarily a contiguous gene version (e.g., macronuclear genes in ciliates [reviewed in reference 27] and mammalian immunoglobulin and T-cell receptor genes [reviewed in reference 22]). The fourth type, previously dubbed “genes in pieces” (5), but here more specifically referred to as “modular genes,” is characterized by discontinuous gene products. Modular mitoribosomal RNAs are relatively frequent, with examples in protists (5, 19, 23, 26, 53, 71) and fungi (20). The few cases of modular mitochondrial protein genes include nad1 (coding for subunit 1 of NADH dehydrogenase) of ciliates (12) and cox2 (coding for cytochrome oxidase subunit 2) of the green alga Scenedesmus obliquus. In all instances, gene modules are encoded on the same DNA molecule, with the notable exception of Scenedesmus cox2, one module of which is encoded by mtDNA (45) and the other by nuclear DNA (21; reviewed in reference 11). Here, we report discontinuous genes of Diplonema mtDNA that differ from all types described above in that each of the multiple gene fragments is located on a distinct mitochondrial chromosome.
Evidently, gene expression of Diplonema cox1 does not involve DNA splicing; otherwise, a contiguous gene should have been detected by PCR. The gene fragments do not code for protein pieces, because the cox1 cDNA is contiguous. Instead, the distinct cox1 gene fragments must be transcribed separately and joined together posttranscriptionally by some kind of trans-splicing. The molecular mechanism by which this process may take place is currently under investigation.
From the existence of at least three separate and dispersed cox1 gene fragments and the failure to amplify genomic cox1 regions of >300 bp, we infer that this gene is split up entirely into ∼250-bp fragments. At an average length of 1,500 to 1,650 bp for euglenozoan cox1 coding sequences (34, 40, 76), the predicted number of cox1 coding fragments in Diplonema should be five or six. With each of these fragments encoded on a separate chromosome and at least two chromosome versions per gene fragment (see Fig. 6B), the genome portion occupied by cox1 alone would then amount to as much as 60 to 84 kbp. This low portion of coding sequence is fully consistent with the absence of detectable genes in the previously determined ∼20 kbp random sequence of Diplonema mtDNA.
Conjectures on structure and size of the Diplonema mtDNA.
Diplonema is one of the few taxa with true monomeric circular mtDNA molecules, together with several animals and kinetoplastids. Otherwise, the shape of mitochondrial chromosomes is generally linear, composed of multiple, tandemly arranged copies, yet appearing circular in physical mapping and sequence assembly (2; reviewed in reference 46). Based on the following estimations, Diplonema may possess one of the largest numbers of mitochondrial chromosomes. Assuming a gene set of 15 genes coded by ∼14 kbp as in T. brucei or Leishmania tarentolae (its closest neighbors with completely sequenced mtDNA; 17), and a single ∼250-bp gene fragment per chromosome, the number of distinct mitochondrial chromosomes in Diplonema would amount to as many as ∼56. This is in stark contrast to the majority of eukaryotes, which possess only a single type of mitochondrial chromosome; notable exceptions are Amoebidium parasiticum (10) and kinetoplastids (reviewed in references 8 and 56). Finally, Diplonema mtDNA might be among the largest mitochondrial genomes. Assuming that there are 56 different chromosomes (see above), each 6 to 7 kbp long, the total size would exceed 360 kbp, which is a size comparable to that of higher plant and ichthyosporean mtDNA (reviewed in reference 23). Obviously, comprehensive mitochondrial genome data will be required to confirm these conjectures.
Evolution of euglenozoan mtDNA structure and expression.
The new data presented here enable several evolutionary issues to be revisited. One is the longstanding controversy over whether the mtDNA network of trypanosomatids is an ancestral or a derived trait in Euglenozoa. The “network-late” scenario is gaining increasing support based on several lines of evidence. First, surveys of mtDNA organization show that bodonid mtDNA circles are not interlocked (reviewed in reference 35); second, protein phylogenies imply that trypanosomatids descended from within the bodonids (59, 61); third, molecular studies indicate that catenated mtDNA is absent from euglenozoan groups diverging prior to kinetoplastids, as shown with E. gracilis (23) and Diplonema (this report; see Fig. 1).
Another contentious question has been which mtDNA conformation is primitive in kinetoplastids, supercoiled as in Parabodo caudatus and Cryptobia helicis, or relaxed circular as in all other kinetoplastids (35; see compilation in Table 1). Our finding of relaxed-circular mtDNA for Diplonema strengthens the notion that this conformation represents the primitive state in Euglenozoa. Finally, the multichromosome mtDNA of kinetoplastids has been commonly considered a recent deviation, but two observations, the numerous mtDNA circles in Diplonema described here and the recently reported highly heterogeneous collection of small DNA molecules recovered from E. gracilis mtDNA, have brought this view into question (23). Therefore, we posit that a multipartite genome structure emerged as early as in the common ancestor of Euglenozoa (see Fig. 1).
What are the implications of this hypothesis for the ancestral gene structure in this lineage? Did euglenozoan mitochondrial genes originally resemble those of kinetoplastids (clustered contiguous cryptogenes plus gRNA genes located on multiple molecules) or, rather, those of Diplonema (discontinuous genes spread over multiple molecules)? Both scenarios are conceivable. The kinetoplastid state could have evolved from a Diplonema-like state by integration of all contiguous, reverse-transcribed mRNAs and rRNAs into a single chromosome and subsequent transformation of the initial gene fragments into gRNA genes. Alternatively, Diplonema-like gene fragments may have originated from compact kinetoplastid-type genes via recombination events, followed by the loss of the contiguous gene versions. Obviously, molecular mtDNA data from a broad taxon diversity are required to shed light on the evolution of gene organization and genome architecture in Euglenozoa.
Outlook.
The gene and genome structure described here are novel and intriguing per se. In addition, these insights enable us to devise new approaches for deciphering the highly repetitive Diplonema mtDNA. Work is in progress to generate comprehensive mitochondrial genome and transcriptome data from Diplonema, which will be instrumental for uncovering the enigmatic mechanism of gene expression in this system.
Acknowledgments
We thank J. Janáček (Institute of Physiology, Prague, Czech Republic), D. Novotná, and P. Masarŏvá (Institute of Parasitology, Cěské Budějovice, Czech Republic) for help with 3D reconstruction; O. Benada (Institute of Microbiology, Prague, Czech Republic) for assistance in electron microscopy of mtDNA; D.A. Maslov and S. Yasuhira (University of California) for kindly providing a D. papillatum genomic clone, subclones, and preliminary sequence data; D. Tremblay for his contribution to mtDNA localization at an early stage of this project; A. G. B. Simpson (Dalhousie University, Halifax, Nova Scotia, Canada) for helpful discussions on mitochondrial ultrastructure; and B. F. Lang, A. Hauth, and L. Koski (Université de Montréal) for helpful discussions on the manuscript.
This research was supported by grants 5022302 and Z60220518 from the Grant Agency of the Czech Academy of Sciences and grant 6007665801 from the Ministry of Education of the Czech Republic to J.L., and a Special Project grant (SP-34) from the Canadian Institutes of Health Research (CIHR), Genetics Program, to G.B. The Canadian Institute for Advanced Research (CIAR), Program for Evolutionary Biology, provided salary and interaction support to G.B. Equipment used in the context of this project was financed by CIHR (MOP-15331) and Genome-Canada/Genome-Quebec (PEP project).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bendich, A. J. 1993. Reaching for the ring: the study of mitochondrial genome structure. Curr. Genet. 24:279-290. [DOI] [PubMed] [Google Scholar]
- 3.Blom, D., A. de Haan, M. van den Berg, P. Sloof, M. Jirků, J. Lukeš, and R. Benne. 1998. RNA editing in the free-living bodonid Bodo saltans. Nucleic Acids Res. 26:1205-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blom, D., A. de Haan, J. van den Burg, M. van den Berg, P. Sloof, M. Jirků, J. Lukeš, and R. Benne. 2000. Mitochondrial minicircles in the free-living bodonid Bodo saltans contain two gRNA gene cassettes and are not found in large networks. RNA 6:121-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boer, P. H., and M. W. Gray. 1988. Scrambled ribosomal RNA gene pieces in Chlamydomonas reinhardtii mitochondrial DNA. Cell 55:399-411. [DOI] [PubMed] [Google Scholar]
- 6.Bonen, L. 1993. Trans-splicing of pre-mRNA in plants, animals, and protists. FASEB J. 7:40-46. [DOI] [PubMed] [Google Scholar]
- 7.Borst, P., F. Fase-Fowler, P. J. Weijers, J. D. Barry, L. Tetley, and K. Vickerman. 1985. Kinetoplast DNA from Trypanosoma vivax and T. congolense. Mol. Biochem. Parasitol. 15:129-142. [DOI] [PubMed] [Google Scholar]
- 8.Borst, P., and A. M. Kroon. 1969. Mitochondrial DNA: physicochemical properties, replication, and genetic function. Int. Rev. Cytol. 26:107-190. [DOI] [PubMed] [Google Scholar]
- 9.Brugerolle, G., J. Lom, E. Nohýnková, and L. Joyon. 1979. Comparaison et évolution des structures cellulaires chez plusieurs espèces de bodonidés et cryptobiidés appartenant aux genres Bodo, Cryptobia et Trypanoplasma (Kinetoplastida, Mastigophora). Protistologica 15:197-221. [Google Scholar]
- 10.Burger, G., L. Forget, Y. Zhu, M. W. Gray, and B. F. Lang. 2003. Unique mitochondrial genome architecture in unicellular relatives of animals. Proc. Natl. Acad. Sci. USA 100:892-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burger, G., M. W. Gray, and B. F. Lang. 2003. Mitochondrial genomes—anything goes. Trends Genet. 19:709-716. [DOI] [PubMed] [Google Scholar]
- 12.Burger, G., Y. Zhu, T. G. Littlejohn, S. J. Greenwood, M. N. Schnare, B. F. Lang, and M. W. Gray. 2000. Complete sequence of the mitochondrial genome of Tetrahymena pyriformis and comparison with Paramecium aurelia mitochondrial DNA. J. Mol. Biol. 297:365-380. [DOI] [PubMed] [Google Scholar]
- 13.Cavalier-Smith, T. 1981. Eukaryote kingdoms: seven or nine? Biosystems 14:461-481. [DOI] [PubMed] [Google Scholar]
- 14.Clayton, D. A., C. A. Smith, J. M. Jordan, M. Teplitz, and J. Vinograd. 1968. Occurrence of complex mitochondrial DNA in normal tissues. Nature 220:976-979. [DOI] [PubMed] [Google Scholar]
- 15.Cowan, R., C. M. Collis, and G. W. Grigg. 1987. Breakage of double-stranded DNA due to single-stranded nicking. J. Theor. Biol. 127:229-245. [DOI] [PubMed] [Google Scholar]
- 16.Dooijes, D., I. Chaves, R. Kieft, A. Dirks-Mulder, W. Martin, and P. Borst. 2000. Base J originally found in Kinetoplastida is also a minor constituent of nuclear DNA of Euglena gracilis. Nucleic Acids Res. 28:3017-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estévez, A. M., and L. Simpson. 1999. Uridine insertion/deletion RNA editing in trypanosome mitochondria—a review. Gene 240:247-260. [DOI] [PubMed] [Google Scholar]
- 18.Farmer, M. A., and R. E. Triemer. 1994. An ultrastructural study of Leptomonas applanatum (Preisig) n.g. (Euglenida). J. Eukaryot. Microbiol. 41:112-119. [Google Scholar]
- 19.Feagin, J. E. 1994. The extrachromosomal DNAs of apicomplexan parasites. Annu. Rev. Microbiol. 48:81-104. [DOI] [PubMed] [Google Scholar]
- 20.Forget, L., J. Ustinova, Z. Wang, V. A. Huss, and B. F. Lang. 2002. Hyaloraphidium curvatum: a linear mitochondrial genome, tRNA editing, and an evolutionary link to lower fungi. Mol. Biol. Evol. 19:310-319. [DOI] [PubMed] [Google Scholar]
- 21.Funes, S., E. Davidson, A. Reyes-Prieto, S. Magallón, P. Herion, M. P. King, and D. González-Halphen. 2002. A green algal apicoplast ancestor. Science 298:2155. [DOI] [PubMed] [Google Scholar]
- 22.Gill, J. I., and M. L. Gulley. 1994. Immunoglobulin and T-cell receptor gene rearrangement. Hematol. Oncol. Clin. N. Am. 8:751-770. [PubMed] [Google Scholar]
- 23.Gray, M. W., B. F. Lang, and G. Burger. 2004. Mitochondria of protists. Annu. Rev. Genet. 38:477-524. [DOI] [PubMed] [Google Scholar]
- 24.Griessman, K. 1914. Ueber marine Flagellaten. Arch. Protistenkd. 32:1-78. [Google Scholar]
- 25.Hajduk, S. L., A. M. Siqueira, and K. Vickerman. 1986. Kinetoplast DNA of Bodo caudatus: a noncatenated structure. Mol. Cell. Biol. 6:4372-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinonen, T. Y., M. N. Schnare, P. G. Young, and M. W. Gray. 1987. Rearranged coding segments, separated by a transfer RNA gene, specify the two parts of a discontinuous large subunit ribosomal RNA in Tetrahymena pyriformis mitochondria. J. Biol. Chem. 262:2879-2887. [PubMed] [Google Scholar]
- 27.Jahn, C. L., and L. A. Klobutcher. 2002. Genome remodeling in ciliated protozoa. Annu. Rev. Microbiol. 56:489-520. [DOI] [PubMed] [Google Scholar]
- 28.Janáček, J., and L. Kubínová. 1998. 3D reconstruction of surfaces captured by confocal microscopy. Acta Stereol. 17:259-264. [Google Scholar]
- 29.Kent, M. L., R. A. Elston, T. A. Nerad, and T. K. Sawyer. 1987. An Isonema-like flagellate (Protozoa: Mastigophora) infection in larval geoduck clams, Panope abrupta. J. Invertebr. Pathol. 50:221-229. [DOI] [PubMed] [Google Scholar]
- 30.Kivic, P. A., and P. L. Walne. 1984. An evaluation of a possible phylogenetic relationship between the Euglenophyta and Kinetoplastida. Origins Life 13:269-288. [Google Scholar]
- 31.Laatsch, T., S. Zauner, B. Stoebe-Maier, K. V. Kowallik, and U.-G. Maier. 2004. Plastid-derived single gene minicircles of the dinoflagellate Ceratium horridum are localized in the nucleus. Mol. Biol. Evol. 21:1318-1322. [DOI] [PubMed] [Google Scholar]
- 32.Larsen, J., and D. J. Patterson. 1990. Some flagellates (Protista) from tropical marine sediments. J. Nat. Hist. 24:801-937. [Google Scholar]
- 33.Liang, X. H., A. Haritan, S. Uliel, and S. Michaeli. 2003. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell 2:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukeš, J., G. J. Arts, J. van den Burg, A. de Haan, F. Opperdoes, P. Sloof, and R. Benne. 1994. Novel pattern of editing regions in mitochondrial transcripts of the cryptobiid Trypanoplasma borreli. EMBO J. 13:5086-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukeš, J., D. L. Guilbride, J. Votýpka, A. Zíková, R. Benne, and P. T. Englund. 2002. Kinetoplast DNA network: evolution of an improbable structure. Eukaryot. Cell 1:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukeš, J., M. Jirků, N. Avliyakulov, and O. Benada. 1998. Pankinetoplast DNA structure in a primitive bodonid flagellate, Cryptobia helicis. EMBO J. 17:838-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madison-Antenucci, S., J. Grams, and S. L. Hajduk. 2002. Editing machines: the complexities of trypanosome RNA editing. Cell 108:435-438. [DOI] [PubMed] [Google Scholar]
- 38.Maslov, D. A., L. Hollar, P. Haghighat, and P. Nawathean. 1998. Demonstration of mRNA editing and localization of guide RNA genes in kinetoplast-mitochondria of the plant trypanosomatid Phytomonas serpens. Mol. Biochem. Parasitol. 93:225-236. [DOI] [PubMed] [Google Scholar]
- 39.Maslov, D. A., and L. Simpson. 1994. RNA editing and mitochondrial genomic organization in the cryptobiid kinetoplastid protozoan Trypanoplasma borreli. Mol. Cell. Biol. 14:8174-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maslov, D. A., S. Yasuhira, and L. Simpson. 1999. Phylogenetic affinities of Diplonema within the Euglenozoa as inferred from the SSU rRNA gene and partial COI protein sequences. Protist 150:33-42. [DOI] [PubMed] [Google Scholar]
- 41.Michel, F., and B. Dujon. 1983. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 2:33-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Montegut-Felkner, A. E., and R. E. Triemer. 1994. Phylogeny of Diplonema ambulator (Larsen and Patterson). I. Homologies of the flagellar apparatus. Eur. J. Protistol. 30:227-237. [Google Scholar]
- 43.Moreira, D., P. Lopéz-Garcia, and K. Vickerman. 2004. An updated view of kinetoplastid phylogeny using environmental sequences and a closer outgroup: proposal for a new classification of the class Kinetoplastea. Int. J. Syst. Evol. Microbiol. 54:1861-1875. [DOI] [PubMed] [Google Scholar]
- 44.Myler, P. J., D. Glick, J. E. Feagin, T. H. Morales, and K. D. Stuart. 1993. Structural organization of the maxicircle variable region of Trypanosoma brucei: identification of potential replication origins and topoisomerase II binding sites. Nucleic Acids Res. 21:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nedelcu, A. M., R. W. Lee, C. Lemieux, M. W. Gray, and G. Burger. 2000. The complete mitochondrial DNA sequence of Scenedesmus obliquus reflects an intermediate stage in the evolution of the green algal mitochondrial genome. Genome Res. 10:819-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nosek, J., and L. Tomáška. 2003. Mitochondrial genome diversity: evolution of the molecular architecture and replication strategy. Curr. Genet. 44:73-84. [DOI] [PubMed] [Google Scholar]
- 47.Patterson, D. J. 1994. Protozoa: evolution and Systematics, p. 1-14. In K. Hausmann and N. Hülsmann (ed.), Progress in protozoology. Gustav Fischer Verlag, Stuttgart, Germany.
- 48.Pearson, W. R. 2000. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol. 132:185-219. [DOI] [PubMed] [Google Scholar]
- 49.Pellegrini, M. 1980. Three-dimensional reconstruction of organelles in Euglena gracilis Z. I. Qualitative and quantitative changes of chloroplasts and mitochondrial reticulum in synchronous photoautotrophic culture. J. Cell Sci. 43:137-166. [DOI] [PubMed] [Google Scholar]
- 50.Philippe, H., P. Lopez, H. Brinkmann, K. Budin, A. Germot, J. Laurent, D. Moreira, M. Muller, and H. Le Guyader. 2000. Early-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions. Proc. R. Soc. Lond. B 267:1213-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pigram, W. J., W. Fuller, and M. E. Davies. 1973. Letter: unwinding the DNA helix by intercalation. J. Mol. Biol. 80:361-365. [DOI] [PubMed] [Google Scholar]
- 52.Porter, D. 1973. Isonema papillatum sp. n., a new colorless marine flagellate: a light- and electron microscopic study. J. Protozool. 20:351-356. [Google Scholar]
- 53.Schnare, M. N., T. Y. Heinonen, P. G. Young, and M. W. Gray. 1986. A discontinuous small subunit ribosomal RNA in Tetrahymena pyriformis mitochondria. J. Biol. Chem. 261:5187-5193. [PubMed] [Google Scholar]
- 54.Schnepf, E. 1994. Light and electron microscopical observations in Rhynchopus coscinodiscivorus spec. nov., a colorless, phagotrophic eugenozoon with concealed flagella. Arch. Protistenkd. 144:63-74. [Google Scholar]
- 55.Schuster, F. L., S. Goldstein, and B. Hershenov. 1968. Ultrastructure of a flagellate, Isonema nigricans nov. gen. nov. sp., from a polluted marine habitat. Protistologica 4:141-149. [Google Scholar]
- 56.Shapiro, T. A., and P. T. Englund. 1995. The structure and replication of kinetoplast DNA. Annu. Rev. Microbiol. 49:117-143. [DOI] [PubMed] [Google Scholar]
- 57.Sharp, P. A. 1987. Trans splicing: variation on a familiar theme? Cell 50:147-148. [DOI] [PubMed] [Google Scholar]
- 58.Simpson, A. G. B. 1997. The identity and composition of the Euglenozoa. Arch. Protistenkd. 148:318-328. [Google Scholar]
- 59.Simpson, A. G. B., E. E. Gill, H. A. Callahan, R. W. Litaker, and A. J. Roger. 2004. Early evolution within kinetoplastids (Euglenozoa), and the late emergence of trypanosomatids. Protist 155:407-422. [DOI] [PubMed] [Google Scholar]
- 60.Simpson, A. G. B., J. Lukeš, and A. J. Roger. 2002. The evolutionary history of kinetoplastids and their kinetoplasts. Mol. Biol. Evol. 19:2071-2083. [DOI] [PubMed] [Google Scholar]
- 61.Simpson, A. G. B., and A. J. Roger. 2004. Protein phylogenies robustly resolve the deep-level relationships within Euglenozoa. Mol. Phylogenet. Evol. 30:201-212. [DOI] [PubMed] [Google Scholar]
- 62.Simpson, L., and R. B. Emeson. 1996. RNA editing. Annu. Rev. Neurosci. 19:27-52. [DOI] [PubMed] [Google Scholar]
- 63.Skuja, H. 1948. Taxonomie des Phytoplanktons einiger Seen in Uppland, Schweden. Symb. Bot. Upsal. 9:5-399. [Google Scholar]
- 64.Sogin, M. L., H. J. Elwood, and J. H. Gunderson. 1986. Evolutionary diversity of eukaryotic small-subunit rRNA genes. Proc. Natl. Acad. Sci. USA 83:1383-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stodola, J. 1967. Encyclopedia of water plants. TFH Publications, Inc., Jersey City, N.J.
- 66.Ŝtolba, P., M. Jirků, and J. Lukeš. 2001. Polykinetoplast DNA structure in Dimastigella trypaniformis and Dimastigella mimosa (Kinetoplastida). Mol. Biochem. Parasitol. 113:323-326. [DOI] [PubMed] [Google Scholar]
- 67.Sturm, N. R., and L. Simpson. 1990. Partially edited mRNAs for cytochrome b and subunit III of cytochrome oxidase from Leishmania tarentolae mitochondria: RNA editing intermediates. Cell 61:871-878. [DOI] [PubMed] [Google Scholar]
- 68.Tessier, L. H., H. van der Speck, J. M. Gualberto, and J. M. Grienenberger. 1997. The cox1 gene from Euglena gracilis: a protist mitochondrial gene without introns and genetic code modifications. Curr. Genet. 31:208-213. [DOI] [PubMed] [Google Scholar]
- 69.Triemer, R. E., and M. A. Farmer. 1991. An ultrastructural comparison of the mitotic apparatus, feeding apparatus, flagellar apparatus and cytoskeleton in euglenoids and kinetoplastids. Protoplasma 164:91-104. [Google Scholar]
- 70.Triemer, R. E., and M. A. Farmer. 1991. An ultrastructural organization of the heterotrophic euglenids and its evolutionary implications, p. 185-204. In D. J. Patterson and J. Larsen (ed.), The biology of free-living heterotrophic flagellates. Clarendon Press, Oxford, United Kingdom
- 71.Turmel, M., C. Lemieux, G. Burger, B. F. Lang, C. Otis, I. Plante, and M. W. Gray. 1999. The complete mitochondrial DNA sequences of Nephroselmis olivacea and Pedinomonas minor. Two radically different evolutionary patterns within green algae. Plant Cell 11:1717-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vickerman, K. 1977. DNA throughout the single mitochondrion of a kinetoplastid flagellate: observations on the ultrastructure of Cryptobia vaginalis. J. Protozool. 24:221-233. [Google Scholar]
- 73.von der Heyden, S., E. E. Chao, K. Vickerman, and T. Cavalier-Smith. 2004. Ribosomal RNA phylogeny of bodonid and diplonemid flagellates and the evolution of euglenozoa. J. Eukaryot. Microbiol. 51:402-416. [DOI] [PubMed] [Google Scholar]
- 74.Xu, C. W., J. C. Hines, M. L. Engel, D. G. Russell, and D. S. Ray. 1996. Nucleus-encoded histone H1-like proteins are associated with kinetoplast DNA in the trypanosomatid Crithidia fasciculata. Mol. Cell. Biol. 16:564-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yasuhira, S., and L. Simpson. 1995. Minicircle-encoded guide RNAs from Crithidia fasciculata. RNA 1:634-643, 1995. [PMC free article] [PubMed] [Google Scholar]
- 76.Yasuhira, S., and L. Simpson. 1997. Phylogenetic affinity of mitochondria of Euglena gracilis and kinetoplastids using cytochrome oxidase I and hsp60. J. Mol. Evol. 44:341-347. [DOI] [PubMed] [Google Scholar]
- 77.Yurchenko, V., R. Hobza, O. Benada, and J. Lukeš. 1999. Trypanosoma avium: large minicircles in the kinetoplast DNA. Exp. Parasitol. 92:215-218. [DOI] [PubMed] [Google Scholar]
- 78.Zíková, A., M. Vancová, M. Jirků, and J. Lukeš. 2003. Cruzella marina (Bodonina, Kinetoplastida): noncatenated structure of poly-kinetoplast DNA. Exp. Parasitol. 104:159-161. [DOI] [PubMed] [Google Scholar]