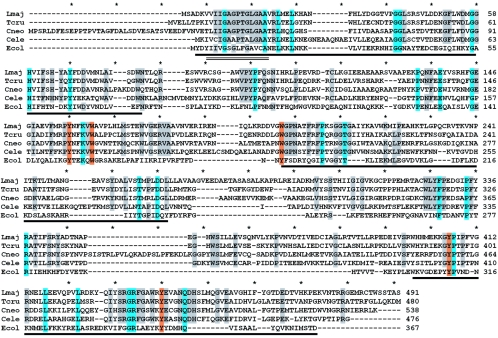
FIG. 1.
Alignment of key eukaryotic and prokaryotic GLF proteins. The predicted proteins encoded by the L. major, T. cruzi, C. neoformans, and C. elegans GLF ORFs were aligned with that of E. coli using methods implemented in the Clustal program (4); the unedited alignment is shown. The amino acid positions are shown on the right, and asterisks mark 10-amino-acid intervals in the alignment. Residues identical in 5/5 sequences are shaded in blue, and residues identical in 4/5 sequences are shaded gray. Residues proposed as part of UDP-Galp binding site are shaded orange (all but one of these are 5/5 identities). The dark underlined regions correspond to the flavin-binding region of E. coli UGM (21); a smaller region showing homology to the Pfam family flavin-containing amine oxidoreductases (PB010804; residues 6 to 19 of the L. major UGM) is underlined by two parallel lines.