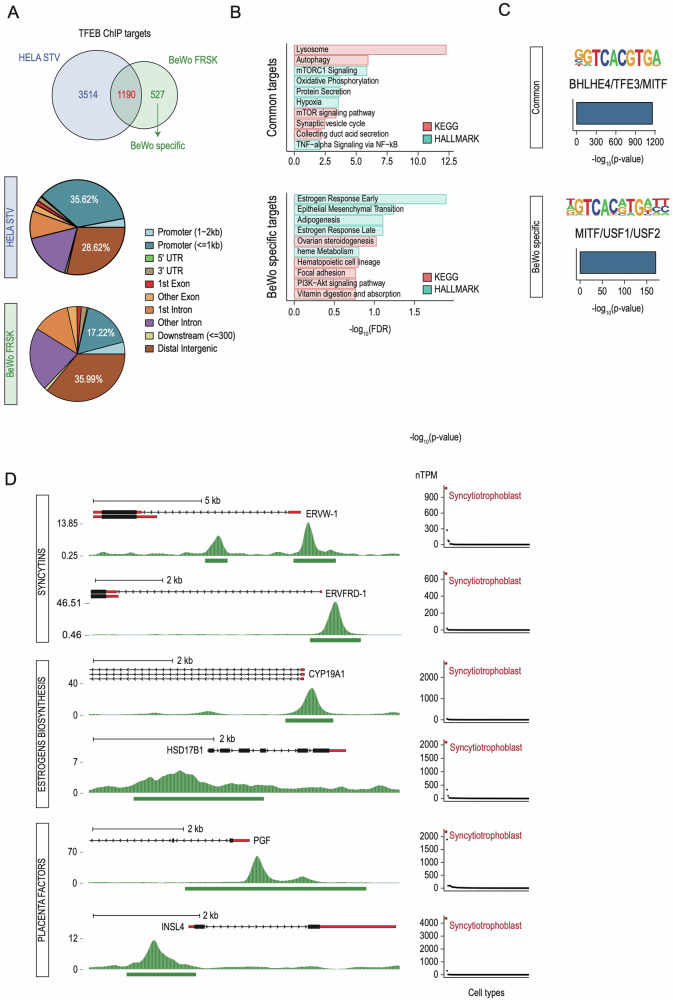
Fig. 2. TFEB targets STB-specific genes.
A (Upper) Venn diagram showing the number of TFEB targets (ChIP peaks at <2 kb from the TSS) in HeLa undergoing starvation (HELA STV) and BeWo cells treated with Forskolin (BeWo FRSK). Common targets (1190) are shown in red and BeWo-specific targets (527) in green. (Lower) Pie charts displaying TFEB binding distribution across the genome in HeLa and BeWo cells at the level of indicated cis-regulatory regions. The distribution of TFEB binding to different regions of target genes, which include promoters (< of 1Kb from the transcriptional start site) and distal intergenic regions, are indicated in the pie charts. B Bar plots of representative term enrichment analysis results using Curated Pathways (KEGG and MSigDB Hallmark collection) of TFEB target genes commonly shared between HeLa and BeWo cells (upper - Common targets) and TFEB BeWo-specific targets (lower - BeWo specific targets). Enriched terms are ranked by −log10 FDR (x-axis). C De-novo motif discovery analysis across the genomic regions displayed in A. For “Common” and “BeWo-specific” categories, the human-specific enriched motif is reported as a motif graph, the best match according to the Homer database, a bar plot displaying statistical significance and the percentage of targets across the genome. D (Left) Representative genome browser snapshots of selected promoters bound by TFEB upon Forskolin treatment in BeWo cells. Both reads distributions as density plots and peak intervals are displayed. Displayed genes are grouped into categories, indicated on the left (“Syncytins” - ERVW-1 and ERVFRD-1; “Estrogen Biosynthesis” - CYP19A1 and HSD17B1; “Placenta Factors” - INSL4 and PGF). (Right) Dot plots indicating the expression levels (nTPM) of indicated TFEB targets across different cell types according to the Human Protein Atlas single-cell datasets. Syncytiotrophoblast is indicated in red.