Fig. 1: The intestine is an environment that harbors a variety of histone acyl marks.
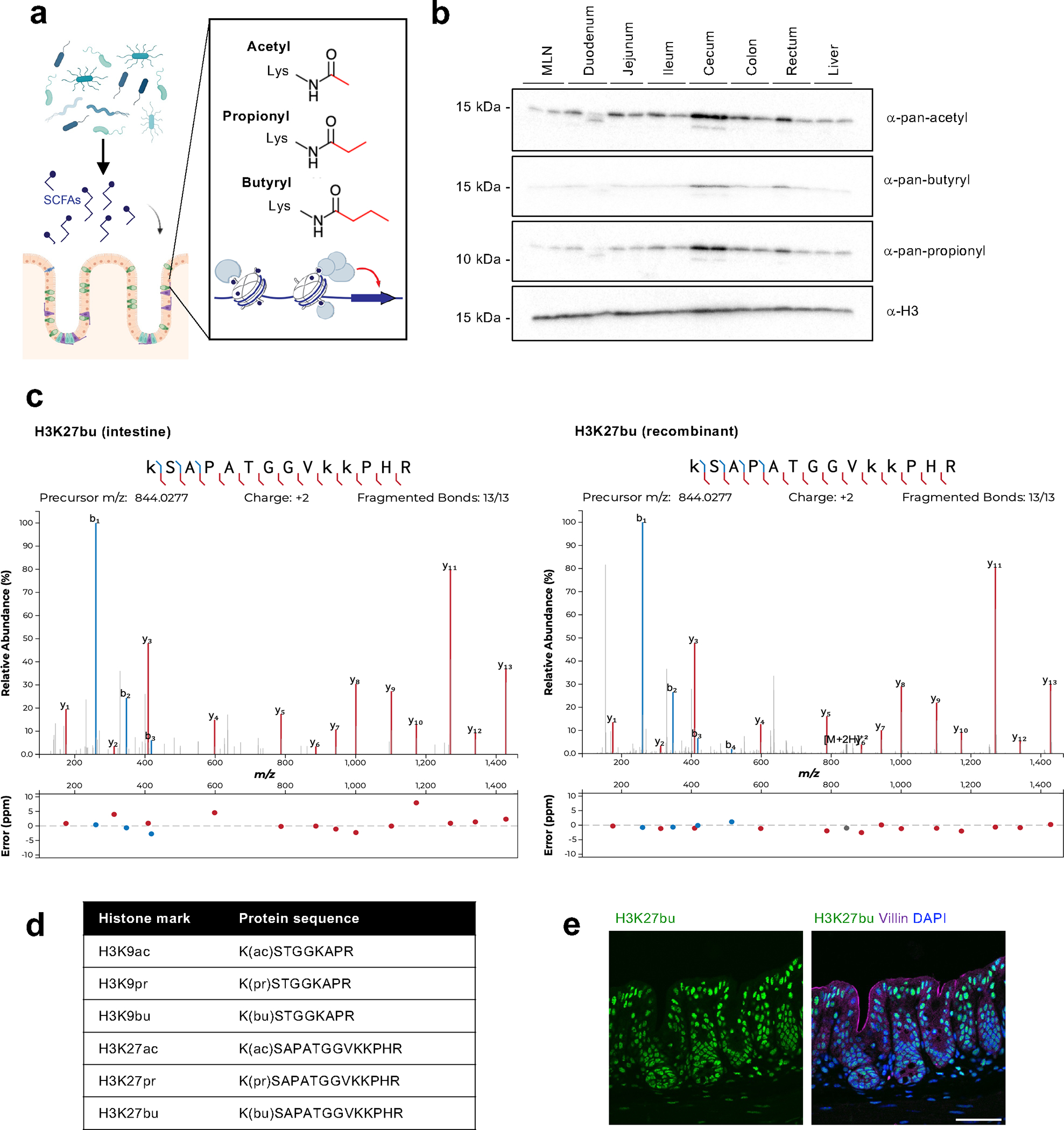
(A) Schematic of overall hypothesis and the structure of select histone acylations. Created with BioRender.com. (B) Tissue distribution of histone acylations. Histones were purified from tissues from female C57BL/6 mice (n = 2 shown) using acid extraction and then subject to immunoblotting with pan-lysine antibodies targeting different acylation modifications. H3 serves as a loading control. (C) Characterization of specific sites of histone acylation. LC-MS/MS analysis of extracted histone from the mouse cecum. Representative fragmentation spectra are shown for endogenous H3K27bu in the intestine (left) and for recombinant H3K27bu on modified nucleosomes (right). (D) Table summarizing the different histone acyl marks detected in the mouse cecum. (E) H3K27bu displays bright staining in the intestinal epithelium. Representative immunofluorescence images of H3K27bu, Villin (marker of epithelium), and DAPI in mouse cecal sections, n = 5 female C57BL/6 mice. Scale bar = 50 μm.
