Fig. 5. LDLR is an alternative B12 uptake receptor.
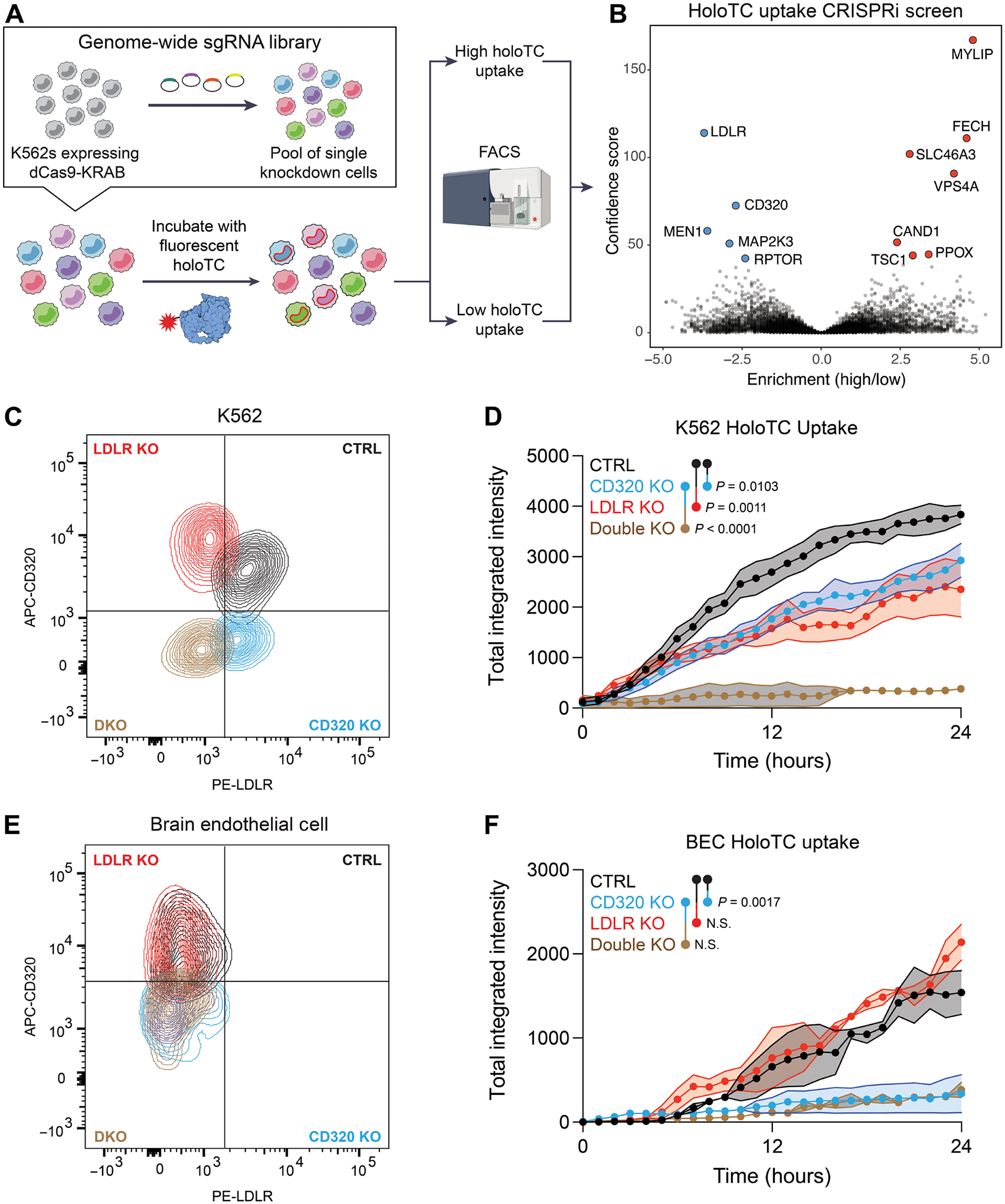
(A) Schematic of a CRISPRi screen for genetic modifiers of holotranscobalamin uptake. K562 cells expressing dCas9-KRAB were infected with a library of sgRNAs targeting the transcriptional start sites of all protein coding genes. This pool of single knockdown cells was then incubated with holotranscobalamin conjugated to a pH-sensitive fluorescent dye. Cells with high (top 5% of red fluorescent signal) and low (bottom 5%) holoTC uptake were separated by FACS and then sequenced and analyzed using casTLE. Screen was performed in technical duplicate. (B) Volcano plot of the genes which when repressed impair (blue) or promote (red) holoTC uptake. The top 12 hits are labeled, meeting an adjusted P value of <0.00001 (Benjamini-Hochberg). (C) Flow cytometry confirmation of control (black), CD320 KO (blue), LDLR KO (red), and double KO (brown) K562 cells. (D) Holotranscobalamin uptake by control (black), CD320 KO (blue), LDLR KO (red), or double KO (brown) K562 cells (n = 3, one-way ANOVA with Tukey’s multiplicity correction, means ± SE). (E) Flow cytometry confirmation of control (black), CD320 KO (blue), LDLR KO (red), and double KO (brown) primary human brain endothelial cells. (F) Holotranscobalamin uptake by control (black), CD320 KO (blue), LDLR KO (red), or double KO (brown) primary brain endothelial cells (n = 2, one-way ANOVA with Tukey’s multiplicity correction, means ± SE). APC, allophycocyanin; PE, phycoerythrin.
