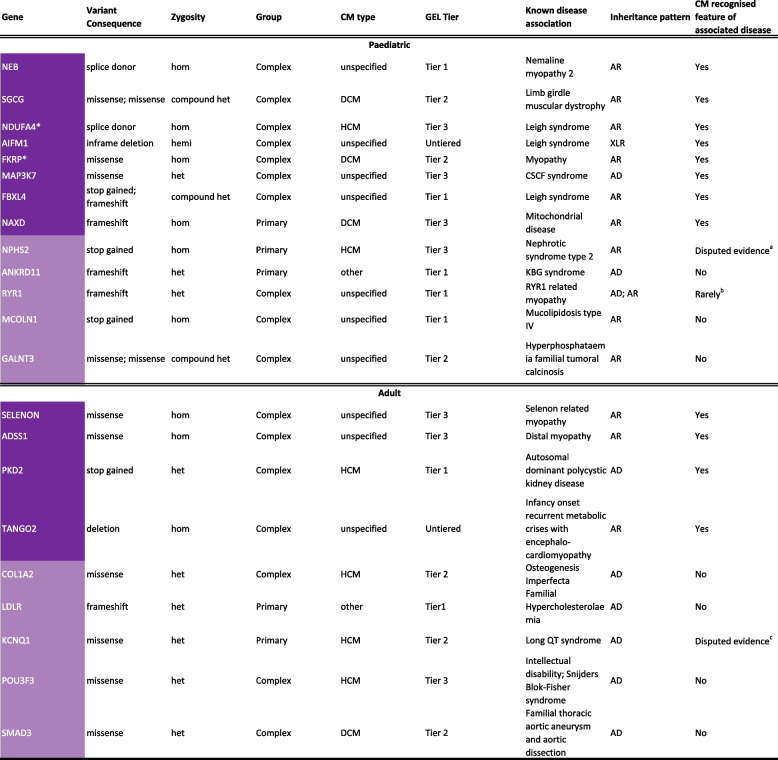
Table 3.
Disease and cardiomyopathy association for twenty-two genes not on the R135 CM panel ‘green’ list where variants were identified and considered diagnostic for cardiomyopathy probands in 100,000 Genomes Project (100KGP)
Left panel - variant details, the phenotype of the proband in 100KGP and how the variant was tiered in GEL's initial analysis
Right panel - known gene disease association, inheritance pattern and whether cardiomyopathy is a recognised feature of the associated disease
Dark purple genes have an associated disease where CM is a recognised feature; light purple genes have an associated disease where CM is not a recognised feature. GEL tier—all variants were tiered on GEL’s initial analysis. Briefly, tier 1: rare protein damaging variants in genes on selected panel(s); tier 2: rare protein altering variants in genes on selected panel(s); tier 3: rare protein altering variants in genes not on selected panel(s). Therefore, if a diagnosis is made in a tier 3 or untiered variant, it suggests the correct panels were not triggered by the participant’s recruitment category or phenotype and the variant(s) was prioritised for another reason, e.g. it was de novo. Additional information about why a variant was prioritised in a particular patient is not always provided in GEL
AD Autosomal dominant, AR Autosomal recessive, CM Cardiomyopathy, CSCF Cardiospondylocarpofacial, GEL Genomics England, het heterozygous, hom homozygous, hemi hemizygous, R135 CM panel, NHS Genomic Medicine Service paediatric or syndromic cardiomyopathy panel (R135 v3.44), XLR X-linked recessive
*FKRP and NDUFA4 are on the R135 CM panel ‘amber’ list (genes with moderate evidence)
aCardiac abnormalities, including left ventricular hypertrophy, have been found in association with the variant p.Arg138* [34], but these findings have not been identified in patients carrying other variants [35]
bCardiac involvement mainly reported in the recessive form; there is a report of adult onset HCM in the dominant form. [36, 37]
cAs per ClinGen, all known HCM cases with KCNQ1 variants also have another variant in, e.g. MYH7 or MYBPC3 that are more likely to be causing the phenotype [38, 39]