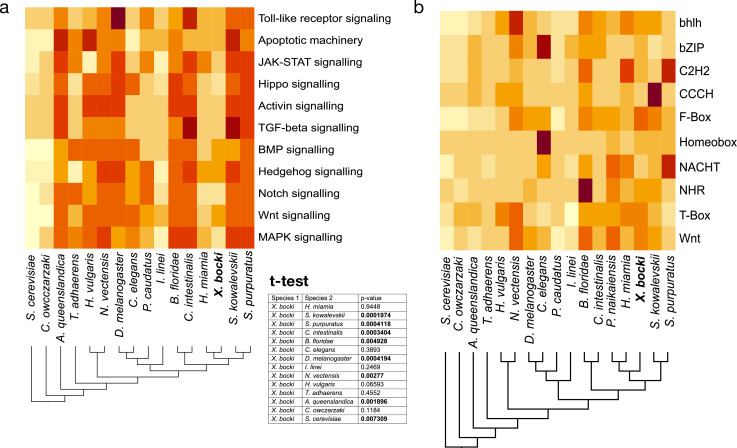
Figure 6. The heatmaps show a comparative measure of relative completeness of signaling pathways based on KEGG and assessed with GenomeMaple or abundance of genes in a given gene-family based on lnterProsScan annotations.
(a) Cell signaling pathways in X. bocki are functionally complete, but in comparison to other species contain less genes. The overall completeness is not significantly different to, for example, the nematode C. elegans (inset, t-test). (b) The number of family members per species in major gene families (based on Pfam domains), like transcription factors, fluctuates in evolution. The X. bocki genome does not appear to contain particularly less or more genes in any of the analyzed families. Due to the comparative nature of the assay, no ‘true’ scale can be given: darker colors indicate higher comparative completeness. Schematic cladograms are drawn by the authors.