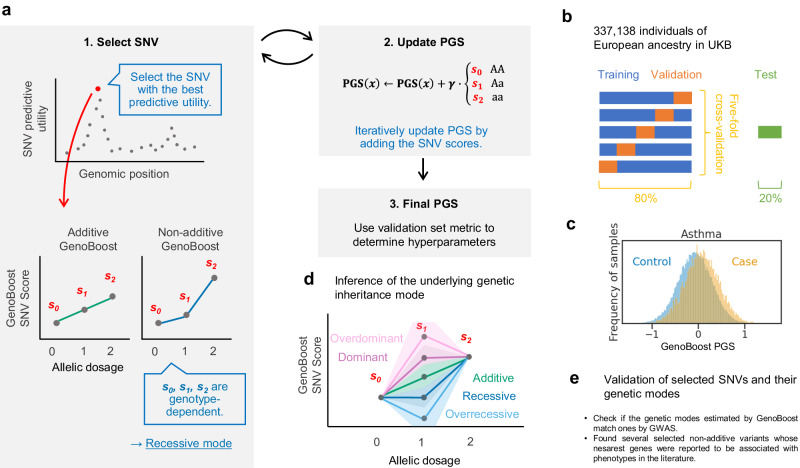
Fig. 1. Schematic overview of the study.
a GenoBoost algorithm fits a polygenic score (PGS) function in an iterative procedure. In each iteration, GenoBoost selects the most informative SNV for trait prediction conditioned on the previously characterized effects and characterizes the genotype-dependent GenoBoost scores, s0, s1, and s2. We considered two types of GenoBoost models: Non-additive GenoBoost and Additive GenoBoost, where an additional constraint among the three GenoBoost scores ensures non-additive genetic dominance effects are always set to zero. GenoBoost iteratively updates its model using two hyperparameters: learning rate γ (0 < γ ≤ 1) and the number of iterations. We optimized the hyperparameters based on the predictive performance in the validation set using five-fold cross-validation. b Model development and evaluation dataset. We randomly split the unrelated white British individuals in UK Biobank into training, validation, and test sets. We applied five-fold cross-validation. c Predictive performance evaluation of PGS models. We used covariate-adjusted pseudo-R2 as the primary metric of predictive performance. d We inferred the mode of inheritance of each genetic variant based on the deviation from the linearity in the three GenoBoost scores (Methods). e We showed that the inferred genetic inheritance is consistent with GWAS-based approaches. We applied GenoBoost to prioritize genetic loci with genetic dominance not previously reported in the GWAS catalog.