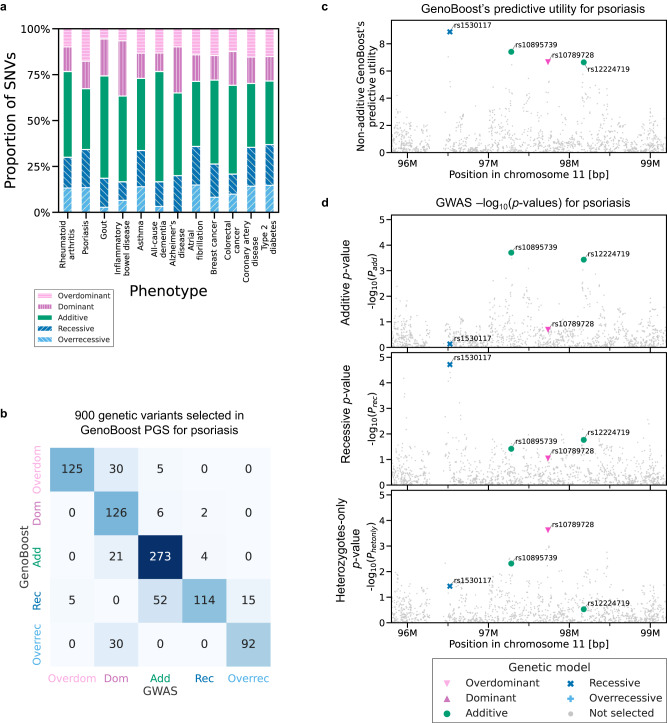
Fig. 3. GenoBoost scores allow for the inference of the mode of inheritance.
a For the genetic variants selected in Non-additive GenoBoost models across the twelve disease outcomes in UK Biobank (x-axis), the fractions of the inferred inheritance mode are shown (y-axis). b We compared the inferred inheritance mode from GenoBoost (y-axis) and the ones from GWAS summary statistics (x-axis) and showed the results as a colored confusion matrix. Add: additive. Dom: dominant. Rec: recessive. c, d Comparison of GenoBoost scores and GWAS p-values, focusing on psoriasis and genetic variants in an intergenic region in chromosome 11. For each genetic variant selected in the Non-additive GenoBoost model within the 3 Mbp window (x-axis), we show the predictive utility of the variant in Non-additive GenoBoost (Supplementary Methods) (c). We also show the statistical significance from GWAS for the variant under additive, recessive, and heterozygous-only regression models (d). The statistical significance is the nominal p-values of the slope of the logistic regression from two-sided tests using up to n = 215,768 samples. Four genetic variants with the largest predictive utilities in GenoBoost are highlighted and colored based on the inferred mode of genetic inheritance. Source data are provided as a Source Data file.