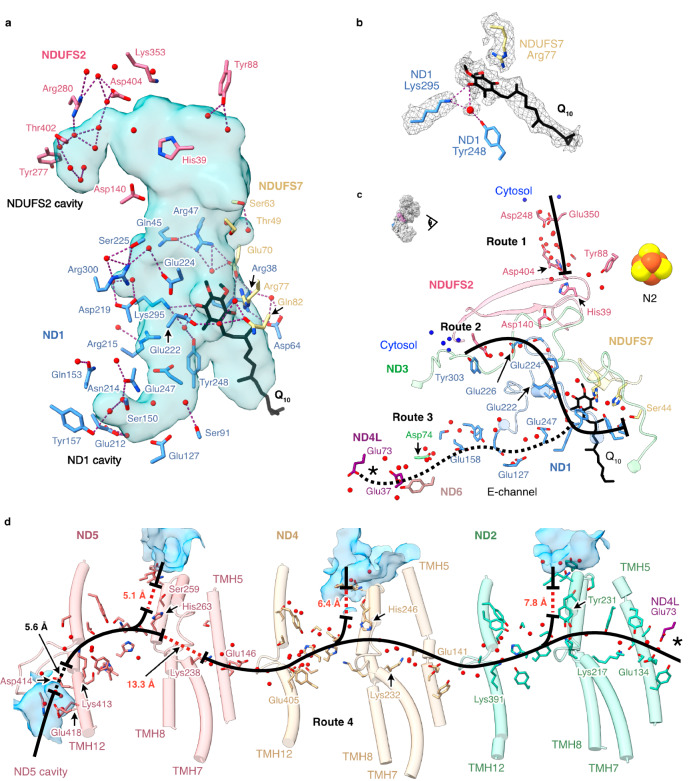
Fig. 6. Hydrogen bonding and Grotthuss competent network of the ubiquinone binding site, E-channel and central axis observed in the turnover-ready resting state of P. denitrificans complex I.
a Hydrogen bonding networks between protonatable residues, water molecules (red spheres) and ubiquinone-10 at the ubiquinone-binding channel identified using the hbonds command in UCSF ChimeraX. The cavity was determined using CASTp analysis with a 1.4 Å radius probe and comprises the substrate binding channel plus an extension into the core of ND1 and an additional lobe in NDUFS2. b Cryo-EM densities for the bound ubiquinone-10, neighbouring protein residues and water molecules. c Grotthuss-competent networks at the ubiquinone-10 binding site (routes 1, 2 and 3), depicting potential ubiquinone-10 protonation routes, and the discontinuous Grotthuss competent network in the E-channel (route 3). d The central axis of ND2, ND4 and ND5 subunits in Pd-CI (route 4) with solvent accessible cavities in blue. In c, d, solid black lines indicate continuous Grotthuss competent networks, while dotted black and red lines denote unobstructed and obstructed gaps in the networks, respectively. Solvent-exposed water molecules are in blue. The asterisks in c and d indicate where the two pathways connect. Detailed views of the Grotthuss networks in c and d are presented in Supplementary Fig. 11a, b, d, e.