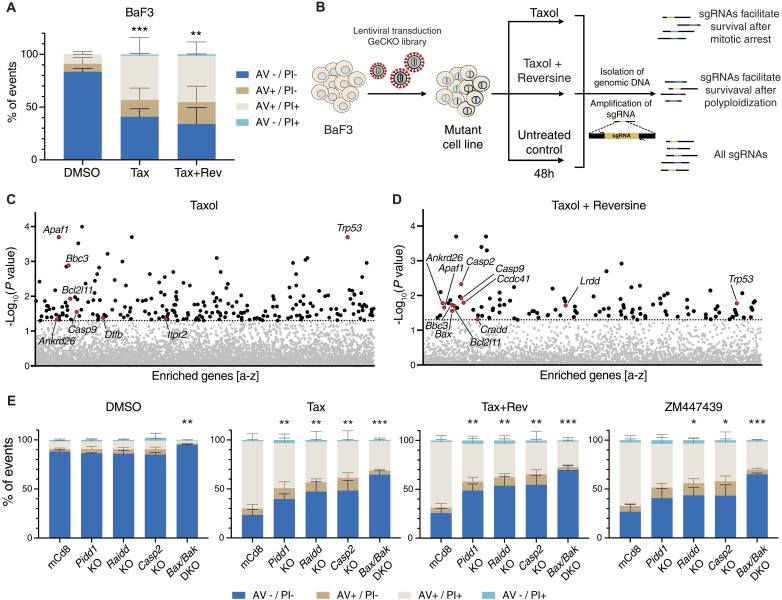
Fig. 1. A CRISPR screen identifies genes involved in the induction of cell death after mitotic perturbation.
(A) BaF3 cells were treated for 48 hours with 50 nM Taxol (Tax) alone or in combination with 500 nM reversine (Tax + Rev) before staining with annexin V (AV) and propidium iodide (PI) followed by flow cytometric analysis. Data are presented as means ± SD of the percentage of events in each staining condition. AV−/PI− = live cells; AV+/PI− = early apoptosis; AV+/PI+ and AV−/PI+ = late apoptosis. N = 3 independent biological replicates. Statistical significance calculated by unpaired t test on the percentage of live cells relative to the DMSO control. (B) Scheme of the CRISPR screen experimental setup. (C) Enriched sgRNAs found in surviving BaF3 cells after Taxol treatment (50 nM). Horizontal dashed line indicates the significance P value cutoff (0.05). See also fig. S2A. (D) Enriched sgRNAs in surviving BaF3 cells after Taxol + reversine treatment (50 nM + 500 nM). Horizontal dashed line indicates the significance P value cutoff (0.05). See also fig. S2B. (E) Annexin V/PI staining and flow cytometric analysis of control (mCd8), Pidd1 KO, Raidd KO, Casp2 KO, and Bak + Bax double KO (DKO) cells, 48 hours after treatment with Taxol (50 nM), Taxol + reversine (50 nM + 500 nM), ZM (2 μM), or DMSO. Data are presented as means ± SD of the percentage of events in each staining condition. N ≥ 3 independent biological replicates. Statistical significance calculated by unpaired t test on the percentage of live cells of each derivative clone relative to the mCd8 control for each drug treatment. See also fig. S2C. *P < 0.05; **P < 0.01; ***P < 0.001.