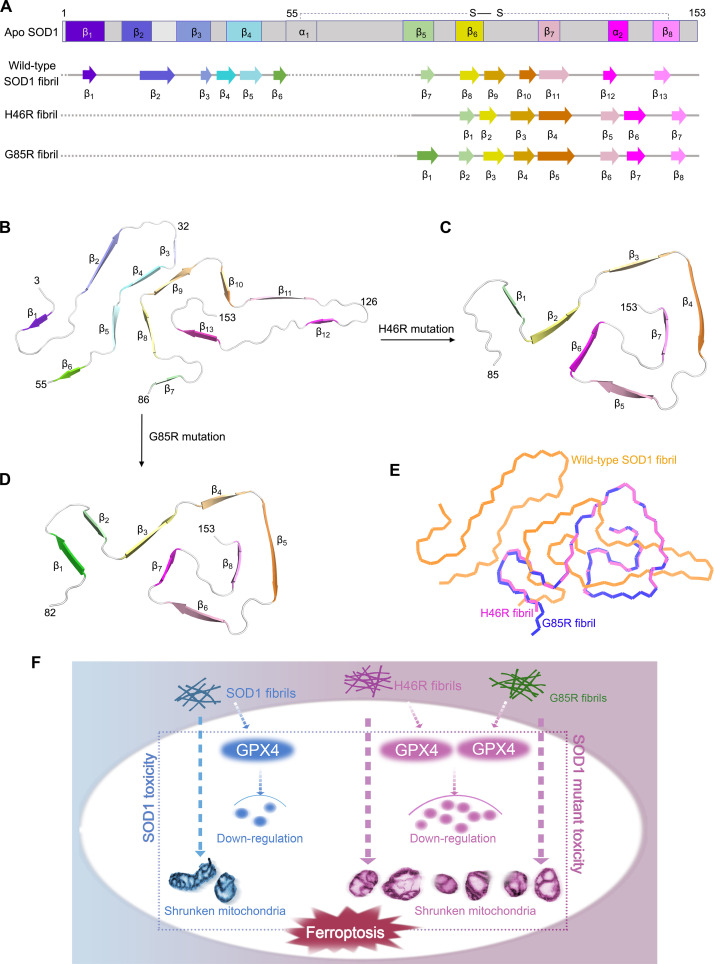
Fig. 7. Comparison of the structures of the apo form of SOD1, the wild-type SOD1 fibril, the H46R fibril, and the G85R fibril.
(A) Sequence alignment of the full-length apo human SOD1 monomer [Protein Data Bank (PDB) 1HL4] (61) with eight β strands (β1 to β8), two α helices, and a single disulfide bond. Sequence alignment of the wild-type SOD1 fibril core comprising residues 3 to 55 and 86 to 153 from the full-length wild-type human SOD1 (PDB 7VZF) (43) with the observed 13 β strands (β1 to β13). Sequence alignment of the H46R fibril core comprising residues 85 to 153 from the full-length human H46R SOD1 with the observed seven β strands colored light green (β1), yellow (β2), gold (β3), orange (β4), pink (β5), magenta (β6), and light magenta (β7) in the C-terminal segment. The dotted line corresponds to residues 1 to 84 not modeled in the cryo-EM density. Sequence alignment of the G85R fibril core comprising residues 82 to 153 from the full-length human G85R SOD1 with the observed eight β strands colored green (β1), light green (β2), yellow (β3), gold (β4), orange (β5), pink (β6), magenta (β7), and light magenta (β8) in the C-terminal segment (bottom). The dotted line corresponds to residues 1 to 81 not modeled in the cryo-EM density. (B to D) Ribbon representation of the structures of a wild-type SOD1 fibril core (B), an H46R fibril core (C), and a G85R fibril core (D), all of which contain one molecular layer and a monomer. (E) Overlay of the structures of a wild-type SOD1 fibril core (orange), an H46R fibril core (magenta), and a G85R fibril core (blue). (F) A hypothetical model shows how fibril seeds from H46R and G85R exhibit higher cytotoxicity and have a significantly higher ability to cause mitochondrial impairment and activate ferroptosis in neuronal cells.