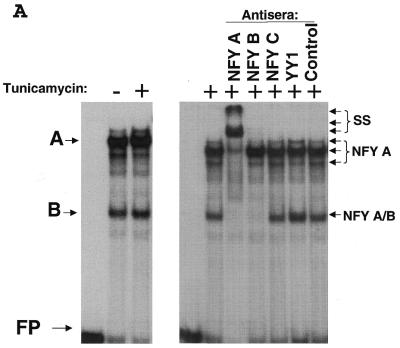
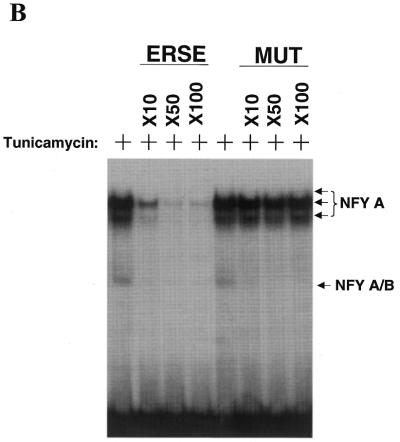
Figure 3.
Identification of NFY as a transcription factor that binds the CHOP ERSE element in both basal and ER-stress stimulated conditions. (A) EMSA experiments performed using nuclear extracts obtained from control (–) and tunicamycin (+) treated NIH 3T3 cells. Nuclear extracts were incubated with the CHOP ERSE in the conditions described in the Materials and Methods section. Results show a similar DNA–protein-binding pattern in both control and tunicamycin-treated conditions. The most prominent complexes are called A and B, respectively. Super-shift experiments using specific antibodies for candidate factors identify the transcription factors NFY-A and NFY-B as the major component of A and B complexes (arrows). Nuclear extracts were incubated in the presence of specific antisera for 15 min prior to adding the DNA probe. (B) Competition experiments using increasing amounts of unlabeled CHOP ERSE (ERSE) and its corresponding mutant ERSE (MUT) oligonucleotides (the sequence of the ERSE and MUT is depicted in Fig. 1C). Unlabeled competitor oligonucleotides were used at 10- to 100-fold molar excess. Extracts are from cells treated with tunicamycin.