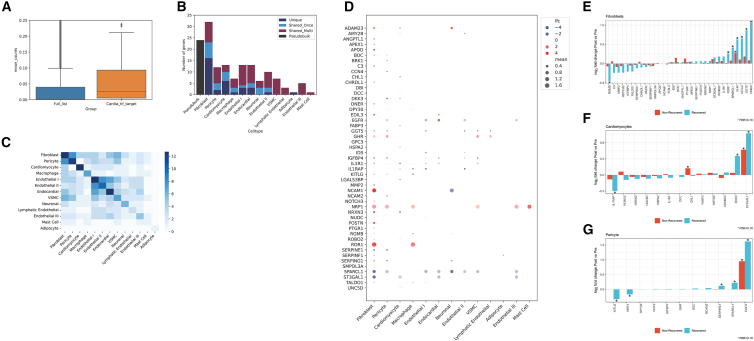
Figure 4.
Single-nucleus myocardial prioritization of clinical targets in human HF
(A) The mean count expression (pseudobulked) between the full list of genes expressed versus those 134 prioritized by our approach, demonstrating a higher myocardial expression for those prioritized genes.
(B) The number of differentially expressed genes by cell type in single-nucleus RNA-seq in HF (dilated cardiomyopathy) versus non-failing (donor) myocardium.18 “Unique” represents genes uniquely differentially expressed in that cell type; “Shared_Once” and “Shared_Multi” represent genes differentially expressed in one other cell type or multiple, respectively. The number of differentially expressed genes in the pseudobulk population is shown in black as a reference and not considered in the shared or unique count for the cell type differentially expressed genes. Fibroblasts, pericytes, and cardiomyocytes are among the top 3 cell types by number of differentially expressed genes.
(C) A heatmap of the number of shared differentially expressed genes across cell types. Fibroblast and pericytes had the most frequent shared differentially expressed genes from our prioritized set.
(D) A heatmap of the log2 fold change for genes differentially expressed in the three prioritized cell types (now visualized across cell types). Genes with blanks signify the absence of reporting of differential expression in the parent data. “lfc” signifies log fold change, and “mean” signifies mean expression.
(E–G) Individual genes across failing and non-failing myocardial samples in fibroblasts, pericytes, and cardiomyocytes by single-nucleus RNA-seq for responders and non-responders across the three cell types. The y axis represents the fold change in individuals who experienced cardiac recovery (more positive = greater expression with hemodynamic unloading with assist device). The asterisks reflect an FDR < 10% (executed only across the displayed transcripts).