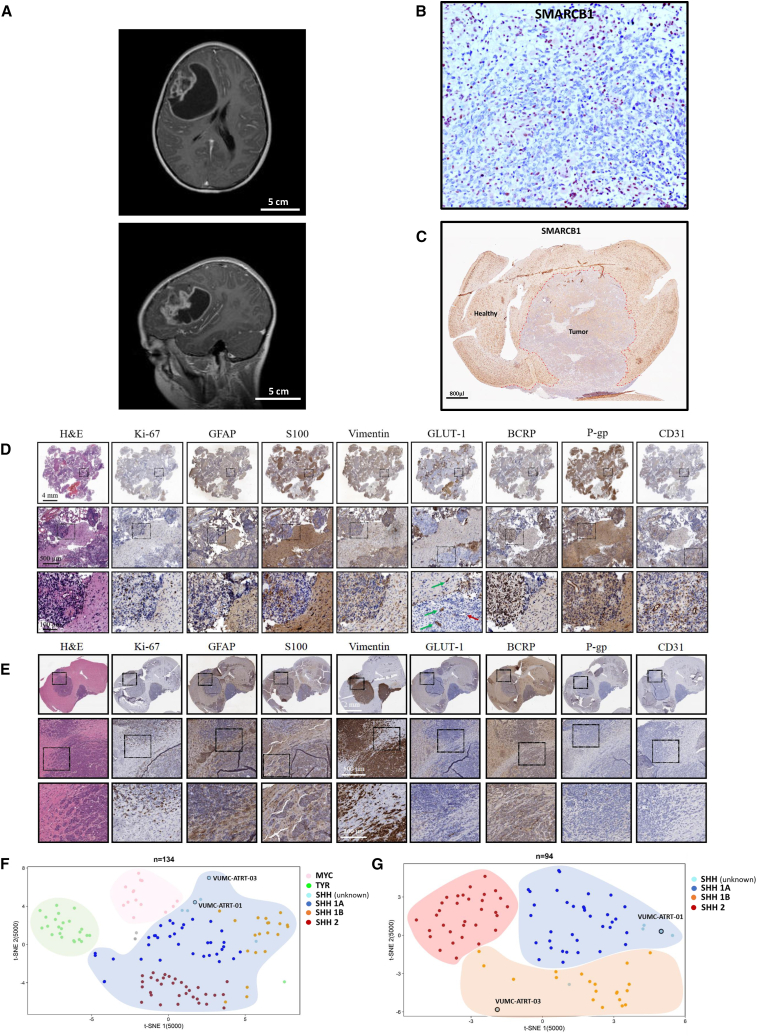
Figure 1.
Establishment of a novel patient-derived SHH-ATRT culture and xenograft model
(A) Diagnostic T2-weighted MRI of the patient from which VUMC-ATRT-03 was derived (top, coronal plane; bottom, sagittal plane).
(B) Immunohistochemistry of patient-derived resection material depicting SMARCB1 expression in brown, revealing typical loss of nuclear SMARCB1 exclusively in tumor cells. The positive nuclei of non-neoplastic and microvascular cells serve as a positive internal control.
(C) Immunohistochemistry for SMARCB1 in a mouse brain carrying a VUMC-ATRT-03 xenograft confirming the absence of SMARCB1 in the nuclei of tumor cells.
(D) Panel of immunohistochemical images depicting ATRT hallmarks in patient-derived resection material (high proliferation: Ki-67; patches of multilineage differentiation: GFAP, S100, vimentin, and keratin; loss of BBB integrity: GLUT-1, BCRP, P-gp, and CD31). Loss of GLUT1 (indicated by the red arrow) indicates intratumor vascular malformations and loss of BBB integrity.
(E) Panel of immunohistochemical images depicting ATRT hallmarks in a mouse brain carrying VUMC-ATRT-03 xenografts (high proliferation: Ki-67; patches of multilineage differentiation: GFAP, S100, vimentin, and keratin; loss of BBB integrity: GLUT-1, BCRP, P-gp, and CD31).
(F) Unsupervised t-distributed stochastic neighbor embedding (t-SNE) clustering of DNA methylation profiles of 134 ATRT samples. Cases are annotated based on previously described subgrouping analysis.9 VUMC-ATRT-01 and VUMC-ATRT-03 methylation profiles annotate to the SHH cluster.
(G) Unsupervised t-SNE clustering of DNA methylation profiles of 94 SHH-ATRT samples. Here, VUMC-ATRT-01 clusters with the SHH-A1 group and the VUMC-ATRT-03 with the SHH-1B group.