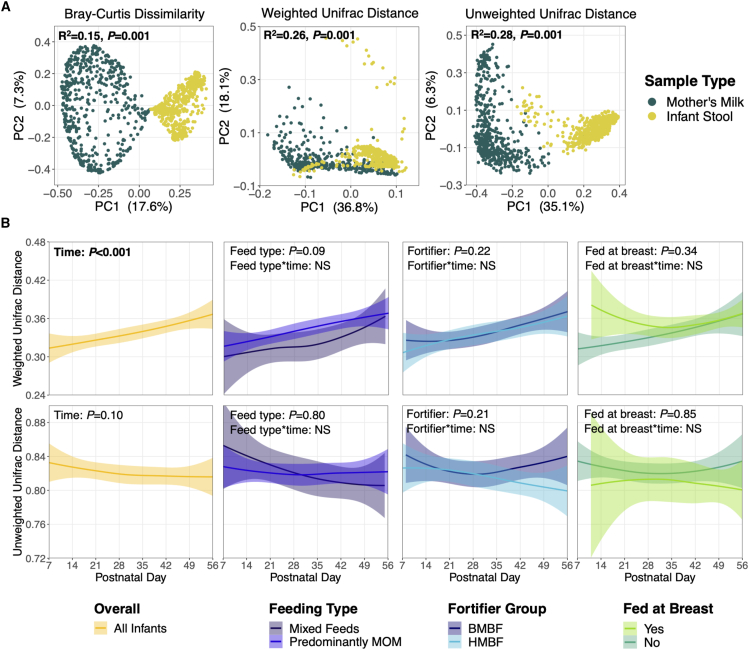
Figure 2.
Microbial beta diversity of paired milk and stool samples
(A) Principal coordinate analysis plots of beta diversity metrics. R2 and p values are from adonis models adjusted for postnatal week, DNA extraction batch, and participant identification. (B) UniFrac distances between paired milk-stool samples were assessed over time for the entire cohort (in yellow) and by feeding variables of interest. Solid lines represent the mean distance between paired samples, while shaded areas represent 95% confidence intervals. p values are from linear mixed-effects models adjusted for postnatal week, DNA extraction batch, infant sex assigned at birth, birth weight stratum, and feeding variables of interest. Interaction terms were tested between the feeding variables and postnatal week but were removed from final models if they were not statistically significant (p > 0.05; denoted with NS). Abbreviations: PC, principal component; MOM, mother’s milk; HMBF, human milk-based fortifier; BMBF, bovine milk-based fortifier.