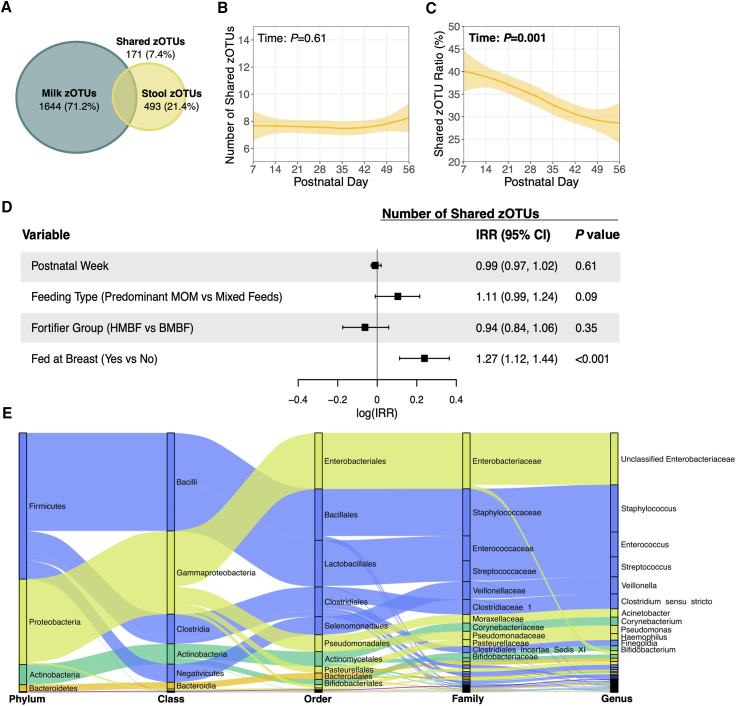
Figure 3.
Shared microbial taxa between paired milk-stool samples
(A) Venn diagram showing the total number of zOTUs unique to milk and stool, and the total number of zOTUs that were shared in paired milk-stool samples. (B) Number of zOTUs shared between paired milk-stool samples over time. Solid lines represent the mean number of shared zOTUs over time, while the shaded areas represent 95% confidence intervals. p values are from linear mixed-effects models adjusted for postnatal week, DNA extraction batch, infant sex assigned at birth, birth weight stratum, and feeding variables of interest. (C) Shared zOTU ratio was calculated as the number of shared zOTUs between paired milk-stool samples divided by the total number of zOTUs in each corresponding stool sample. This model was adjusted as described in (B). (D) Relationships between postnatal week, feeding variables of interest, and the likelihood of sharing a greater number of zOTUs between milk-stool pairings were assessed using an adjusted repeated measures Poisson regression model as described in (B). (E) Alluvial diagram depicting the taxonomy of shared zOTUs between paired milk-stool samples. Node sizes represent the number of milk-stool pairings that shared a zOTU mapping back to the specified taxa. For visual clarity, only taxa that were shared in approximately 10% of milk-stool pairings are listed. Abbreviations: zOTUs, zero-radius operational taxonomic units; IRR, incidence rate ratio; MOM, mother’s milk; HMBF, human milk-based fortifier; BMBF, bovine milk-based fortifier.