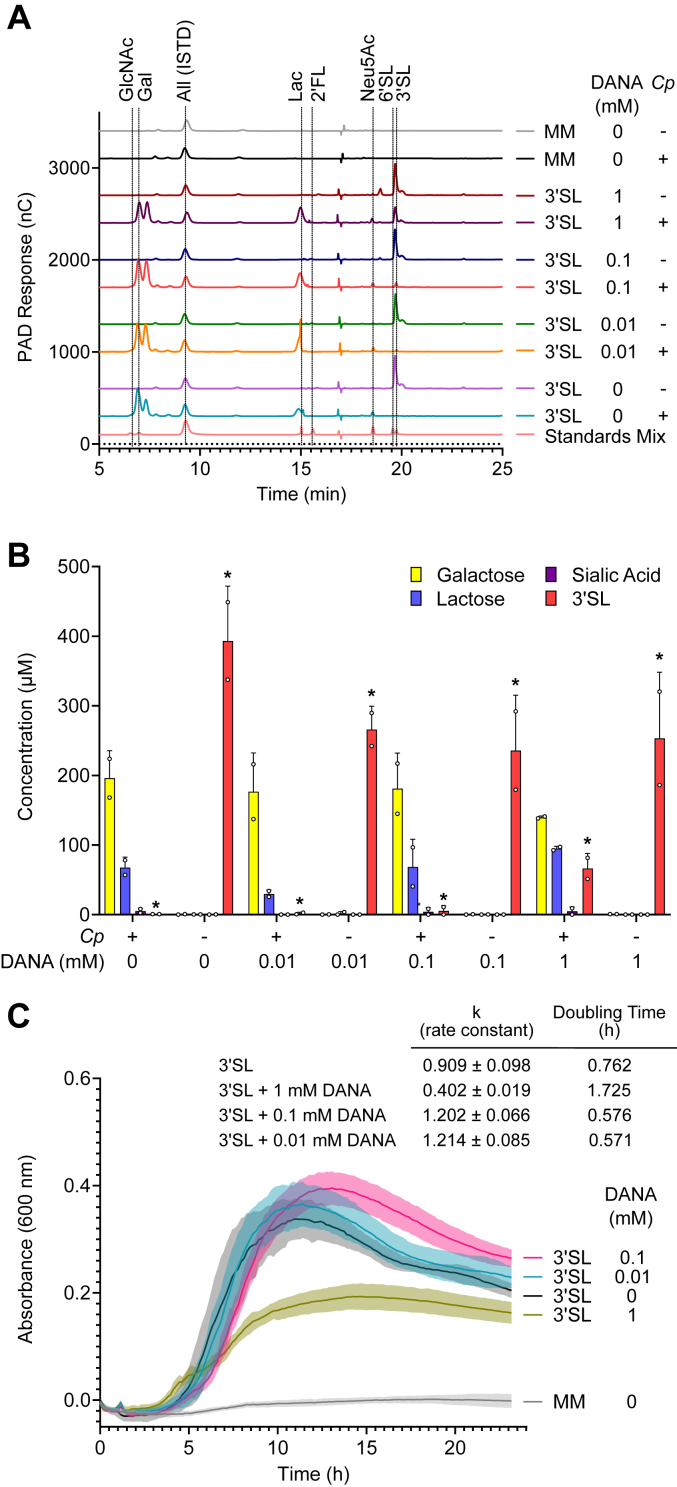
Figure 9.
Detection of carbohydrates remaining in Clostridium perfringens culture supernatants.A, representative HPAEC-PAD traces of filtered cell-free supernatants from 24 h cultures or negative controls containing 0.5% 3′-sialyllactose (3′-SL) and indicated concentrations of DANA. A dilution series of a standards mix of known milk oligosaccharides and monosaccharides was run in parallel with the samples to allow for quantification with an allose internal standard (ISTD) to account for variation in instrument injection volumes. Peaks from standards in the mix are indicated. B, quantification of known carbohydrates from C. perfringens supernatants. Error bars show the standard deviation of the mean (SD; n = 2 biological replicates) and asterisks indicate significant (p < 0.05) differences relative to the leftmost treatment group (+C. perfringens, - DANA) using a 2-way ANOVA. C, Clostridium perfringens growth curves in the presence of increasing concentrations of DANA neuraminidase inhibitor using 3′-SL as a substrate, with optical density monitored by absorbance quantitation at 600 nm. Averages of six growth curves are shown, with areas representing the standard deviation (SD). MM indicates minimalized medium with no carbohydrate. Bacterial growth kinetics were calculated based on exponential growth (GraphPad Prism), and rate constants and doubling times are listed for each growth condition. DANA, N-acetyl-2,3-dehydro-2-deoxyneuraminic acid; HPAEC-PAD, high-performance anion-exchange chromatography with pulsed amperometric detection.