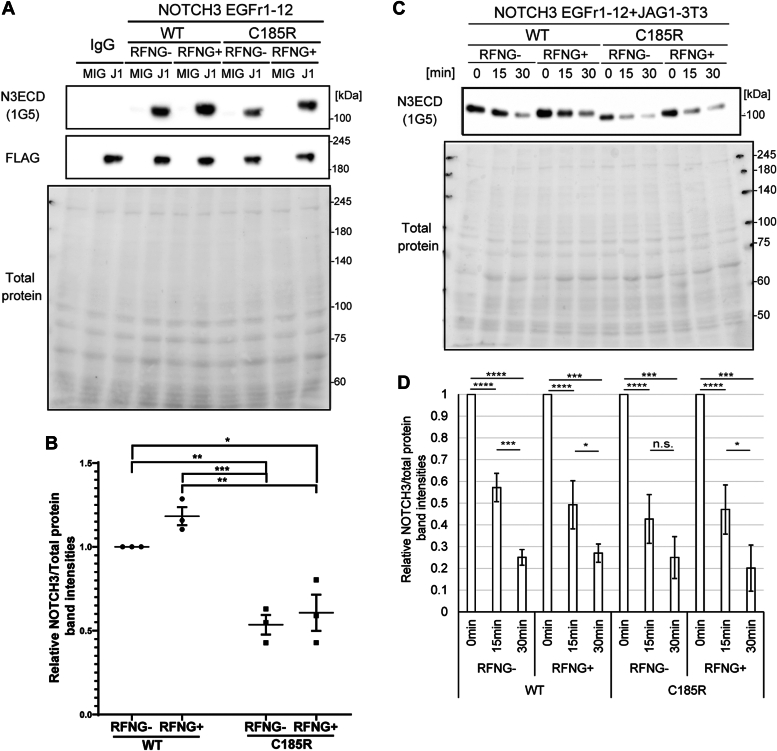
Figure 4.
RFNG does not affect JAG1-mediated degradation of preclustered NOTCH3 WT or C185R soluble proteins.A, representative Western blot of lysates collected from MIG-3T3 and JAG1-3T3 cells after binding to purified NOTCH3 EGFr1-12 WT or C185R, with or without RFNG. NOTCH3 and JAG1 were immunoblotted with antibodies against NOTCH3ECD and FLAG. Ponceau stained total protein. B, relative amount of NOTCH3 EGFr1-12 WT and C185R binding to JAG1, as determined by the blots shown in (A) (independent biological replicates N = 3). The values were calculated by normalizing the relative NOTCH3 band intensities to those of the NOTCH3 WT without RFNG (WT RFNG-). C, representative blot of lysates collected from JAG1-3T3 cells after taking up purified NOTCH3 EGFr1-12 WT and C185R, with or without RFNG-mediated modification. D, relative amounts of NOTCH3 EGFr1-12 WT and C185R after degradation by JAG1, as determined by the blots shown in (C) (independent biological replicates N = 3). The NOTCH3 band intensities were divided by the total protein band intensities. The values are shown as NOTCH3 band intensities relative to those at 0 min. Data information: In B and D, data are presented as mean ± SDs. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 (B: Tukey’s post hoc test following two-way ANOVA; C: Tukey’s post hoc test following one-way ANOVA). EGFr, EGF repeat; JAG1, Jagged 1; RFNG, Radical fringe.