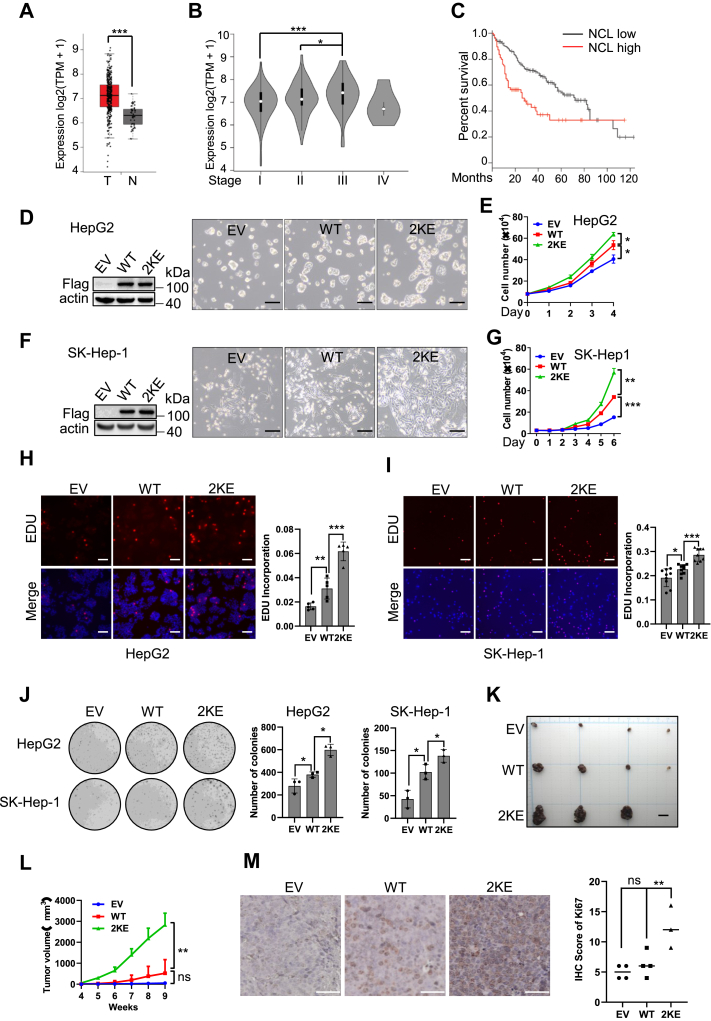
Figure 3.
NCL malonylation promotes HCC cell proliferation in vitro and in vivo.A, comparative analysis of NCL mRNA expression between liver tumor specimens and the normal tissues was performed. The mRNA data was derived from the cancer genome atlas (TCGA) database and analyzed by GEPIA2. n(T) = 369, n(N) = 160. B, comparative analysis of NCL expression between different stages of liver cancer. Stage I, n = 168; stage II, n = 84; stage III, n = 82; stage IV, n = 6. C, Kaplan-Meier survival curve of patients with low (n = 91) or high (n = 91) level expression of NCL. (p = 0.00046, Log-rank test). The quartile cutpoint is used to separate the low (below the 25th percentile) and high (above the 75th percentile) groups. D–I, HepG2 cells and SK-Hep1 cells were infected with lentivirus containing EV, WT NCL (NCLWT), or malonylation mimicking mutant (NCL2KE), followed by selection with puromycin. Polyclonal stable cells were used for cell proliferation assay. Representative pictures showing the confluence (24 h after seeding) of HepG2 (D) and SK-Hep1 cells (F); scale bar indicating 200 μm. E, cell growth curve for HepG2 cells within 96 h. G, cell growth curve for SK-Hep1 cells within 144 h. (H) HepG2 and (I) SK-Hep-1 stable cells were stained with EDU (red) and Hoechst (blue). Representative pictures were shown on the left panel. Scale bar indicates 100 μm. Right panel showing the bar graph in which EDU signal was quantified and normalized by Hoechst signal intensity. J, colony formation ability of HepG2 and SK-Hep-1 cells with stable expression of the indicated ectopic protein. Left panel: representative pictures; right panel showing the quantification of colonies. K–M, HepG2 cells with stable expression of the indicated ectopic protein were subcutaneously injected into nude mice to establish HCC xenograft model. Nine weeks later, mice were sacrificed. K, image of xenograft tumors. Scale bar indicates 10 mm. L, tumor volumes were measured and recorded every week. M, Ki-67 staining in xenograft tumors. Left panel: representative images of Ki-67 staining, scale bar indicating 50 μm; right panel: quantification of Ki-67 staining. ∗p < 0.05,∗∗∗p < 0.001,∗∗∗∗p < 0.0001, n = 3. Error bars represent ± SD.