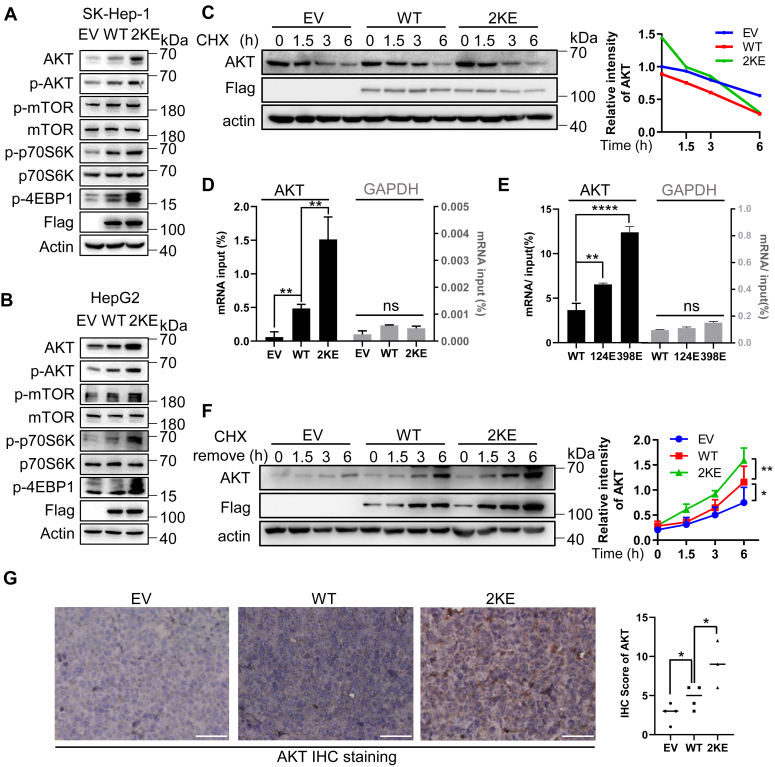
Figure 5.
Malonylated NCL binds to AKT mRNA and facilitates AKT translation.A–B, protein expression of the PI3K/AKT/mTOR pathway in SK-Hep1(A) and HepG2(B) stable cells. Total protein of each cell line was processed by immunoblotting using antibodies against AKT, p-AKT, mTOR, p-mTOR, P70S6K1, p-P70S6K1, p-4E-BP1. Protein loading was normalized with actin. C, half-life of AKT protein in EV, NCLWT, and NCL2KE stable SK-Hep1 cells. The indicated SK-Hep1 cells were treated with 100 μg/ml cycloheximide (CHX) for the time-course as indicated. Total cell lysates were collected for immunoblotting of AKT, Flag, and actin. D–E, RNA-IP showing the binding affinity of indicated ectopic NCL to AKT mRNA. Cell lysates from EV, NCLWT, and NCL2KE (D) or NCLWT, NCLK124E, and NCLK398E (E) stable SK-Hep-1 cells were immunoprecipitated with anti-Flag antibody. The co-precipitated AKT or GAPDH (included as a negative control) mRNA was quantified using RT-qPCR. Results (the mean ± SD, n = 3) were presented as percentages of IP signal/input signal (% input). F, EV, NCLWT, and NCL2KE stable SK-Hep-1 cells were pretreated with 100 μg/ml cycloheximide for 12 h. Cycloheximide was then washed out, and cells were incubated for the time course as indicated. AKT synthesis levels were detected by immunoblotting. Right panel: AKT expression was quantified by ImageJ, with each point representing the mean ± SD. G, histological analysis of AKT expression in HCC xenografts. HepG2 xenograft mice with indicated expression of NCLWT and NCL2KE were established as described above. Left panel: representative images of AKT IHC staining; right penal: AKT staining was quantified and the scores were presented in the bar graph. Scale bar indicates 50 μm. Each point represents the mean ± SD, ns: not significant, ∗p < 0.01, ∗∗p < 0.01, ∗∗∗p < 0.001.