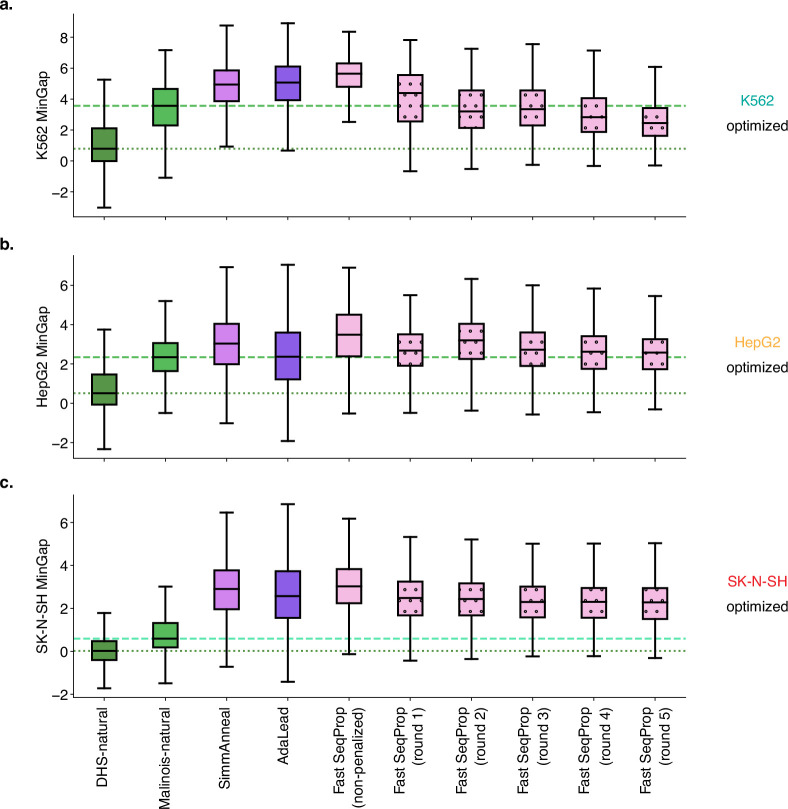
Extended Data Fig. 6. Library MinGap.
(a) Malinois improves identification of CREs with K562-specific activity and synthetic sequence generation enables creation of CREs with enhanced functions. Distribution of MPRA-measured K562-specific activity in various candidate CRE groups. Green and aquamarine lines indicate median MinGap of DHS-natural and Malinois-natural candidates respectively. Sequences with a replicate log2FC standard error greater than 1 in any cell type were omitted from the plots. Boxes demarcate the 25th, 50th, and 75th percentile values, while whiskers indicate the outermost point within 1.5 times the interquartile range from the edges of the boxes. Number of sequences left to right n = 3,729; 3,410; 3,584; 3,545; 3,738; 955; 958; 967; 958; 962. (b) Same as (a) except quantification of candidate sequences targeting HepG2. Number of sequences left to right n = 3,757; 3,727; 3,703; 3,531; 3,683; 917; 938; 961; 953; 966. (c) Same as (a) except quantification of candidate sequences targeting SK-N-SH. Number of sequences left to right n = 3,261; 3,804; 3,894; 3,868; 3,915; 978; 968; 976; 972; 972.