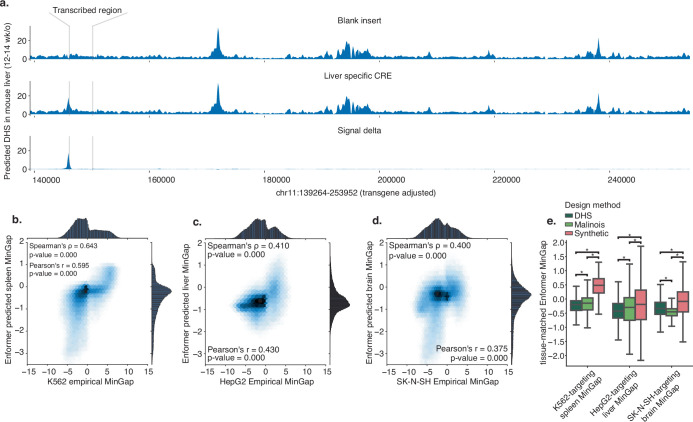
Extended Data Fig. 8. Enformer based prioritization of oligos for in vivo tests.
(a) Enformer can predict CRE-driven changes in epigenetic and transcription dynamics of transgenes inserted into the H11 safe harbour locus in mice. Three example sequence tracks display predicted DHS signals observed in the livers of 15.5 day old mice. Transgene transcription start site and poly-adenylation signal are indicated by the grey bars. The first track is the predicted signal when the input sequence at the CRE insertion site is all Ns. The second track is an example predicting using a validated HepG2-specific synthetic CRE. The third displays the differential DHS effect. (b) Empirical K562 MinGap measurements are well correlated with Enformer-predicted features of spleen-specific transcriptional activation (Methods). (c) Empirical HepG2 MinGap measurements are also well correlated with Enformer-predicted features of liver-specific transcriptional activation. (d) Empirical SK-N-SH MinGap measurements are also well correlated with Enfomer-predicted features of neural-specific transcriptional activation. (e) Enformer-based cell type matched tissue-specific transcriptional activation predictions (K562 matched to spleen, HepG2 matched to liver, SK-N-SH matched to adult brain). Stars indicate family-wise error rate corrected p-values < 1e-4 (In each trio of boxes, n = 4,000; 4,000; 12,000 elements for the DHS, Malinois, and synthetic groups, respectively).