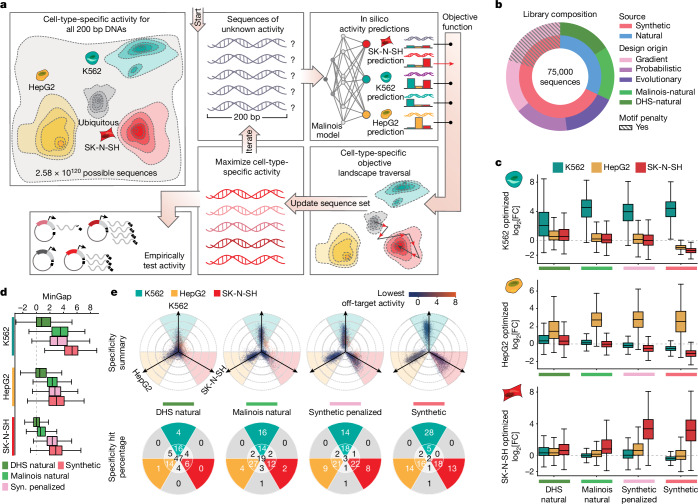
Fig. 2. CODA effectively designs cell-type-specific CREs.
a, CODA designs synthetic elements by iteratively updating sequences to improve predicted function. Malinois predicts CRE activity and an objective function directs sequence updates. After a stopping criteria is met, candidates are nominated for experimental validation. b, The MPRA library composition used to empirically evaluate candidate CREs. c, The distribution of MPRA log2[FC] measurements, each row of the boxes corresponds to candidate CREs intended to drive specific expression in K562, HepG2 and SK-N-SH cells, respectively. Each box indicates measurements in either K562 (teal), HepG2 (yellow) or SK-N-SH (red) cells for the set of sequences nominated by the indicated design strategy on the x axis (left to right, top to bottom, n = 3,729; 3,410; 4,800; 10,867; 3,757; 3,727; 4,735; 10,917; 3,261; 3,804; 4,866; 11,677 elements). d, The distribution of MinGap scores (box plots), quantifying specificity; the colour indicates the intended target cell type (K562, teal; HepG2, yellow; SK-N-SH, red; n values are as described in c, ordered from top to bottom). For the box plots in c and d, the centre line shows the 50th percentile, the box limits show the 25th and 75th percentiles, and the whiskers indicate the outermost point within 1.5× the interquartile range from the edges of the boxes. Syn. indicates synthetic. e, Propeller plots for each sequence group (top). The radial distance corresponds to the distance between the maximum and minimum cell type activity values, and the angle of deviation from an axis quantifies the relative activity of the highest off-target cell type (Methods). The dot colours indicate minimum activity across cell types. Bottom, the percentages of points in each delimited area rounded to the nearest integer. The groups synthetic and synthetic-penalized were randomly subsampled to resemble the size of the two natural groups. From left to right, n = 10,747; 10,941; 12,000 and 12,000 (Supplementary Fig. 8).