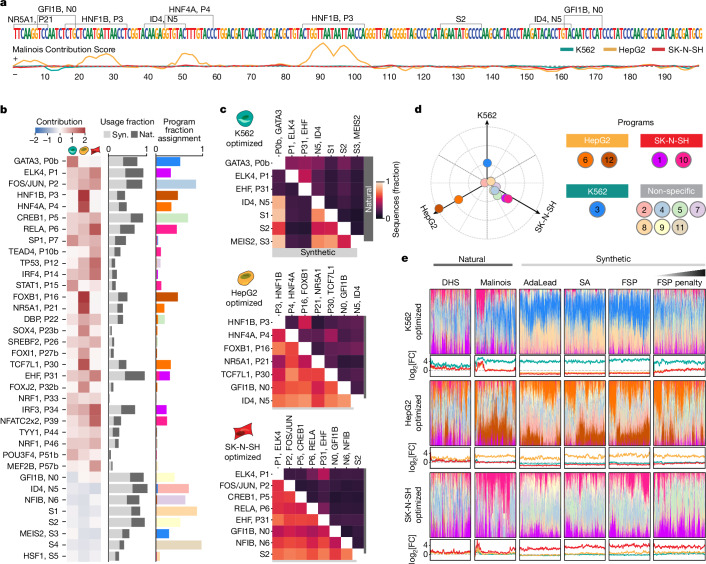
Fig. 3. Interpreting functional sequence content.
a, Malinois contribution scores of a representative synthetic CRE designed to drive HepG2-cell-specific expression. Enriched motifs are demarcated above the sequence and contribution scores are plotted below (K562, teal; HepG2, yellow; SK-N-SH, red) (Methods). b, The average contributions of core motifs in K562, HepG2 and SK-N-SH cells (left to right columns) (left). Middle, motif enrichment in synthetic (light grey) and natural (dark grey) sequences. The x axis represents fraction of sequences in each group containing the motif denoted on the y axis. Right, motif program association derived from the NMF feature matrix. The colours correspond to programs listed in d. c, Co-occurrences of enriched motifs. The colour indicates the percentage of sequences in each group containing a pair of motifs (Methods and Supplementary Fig. 13). The upper and lower triangular percentages correspond to natural and synthetic sequences, respectively. d, The empirical program function was calculated using a weighted average of MPRA log2[FC] scores based on program mixture displayed in e. Ten specificity-driving programs were identified using the same criteria applied to sequences (bright coloured points). Seven programs are not associated with cell-type-specific transcription (pastel colours). Program 11 is overplotted by program 8, and program 4 partially obstructs program 9 on the plot. e, NMF decomposition of synthetic and natural sequences based on enriched motif content. For each sequence, programs are coloured based on the key in d and are plotted as a fraction of the total program content. Sequences not assigned to any program with any frequency yield a blank bar. Line plots display empirical activity in K562 (teal), HepG2 (yellow) and SK-N-SH (red) cells. SA, simulated annealing; FSP, Fast SeqProp. Sequences in each subpanel are sorted by hierarchical clustering based on program content (FSP penalty, n = 5,000; all others, n = 4,000).