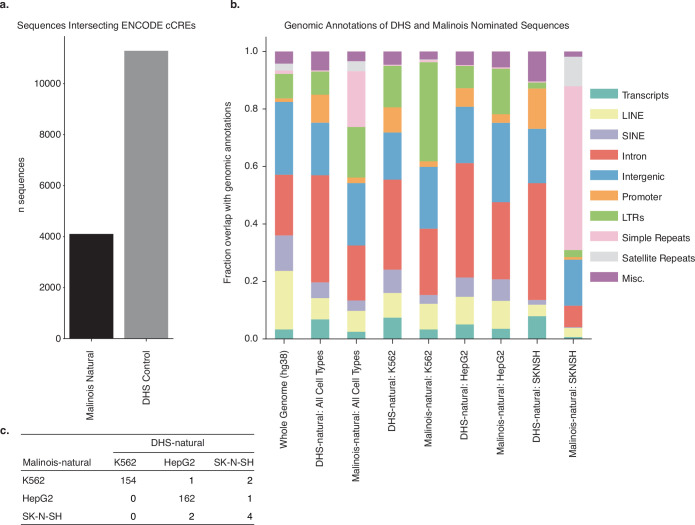
Extended Data Fig. 3. Annotation of naturally occurring sequences.
(a) Sequences nominated by DHS accessibility (DHS-natural) and by Malinois (Malinois-natural) were intersected with ENCODE cCREs (promoter-like sequences, proximal enhancer-like sequences, distal enhancer-like sequences, and CTCF-only) to determine overlap with existing putative regulatory elements. 94% of DHS-natural sequences intersect a cCRE while only 34.2% of Malinois-natural sequences intersect a cCRE suggesting that Malinois may exploit sequences features not captured by typical cCRE measures to select a sequence that drives cell type-specific activity. (b) To explore additional genomic features that may overlap DHS-natural and Malinois-natural sequences were annotated using annotatePeaks.pl from the HOMER suite. Annotations were generated for the whole genome (hg38), the DHS-natural and Malinois-natural libraries as a whole, as well as DHS-natural and Malinois-natural by individual cell type. DHS-natural and Malinois-natural largely resemble the distribution of annotations genome-wide barring an overrepresentation of simple repeats in Malinois-natural sequences driven by SK-N-SH sequences. Despite this, selected sequences seem to be a representative sample of genomic features. (c) DHS-natural and Malinois-natural sequences were intersected to determine overlap between naturally occurring sequences. Notably overlap was minimal between selection methods (0.10%-4.1%) depending on cell type.