Extended Data Fig. 5. MPRA of mutated sequences.
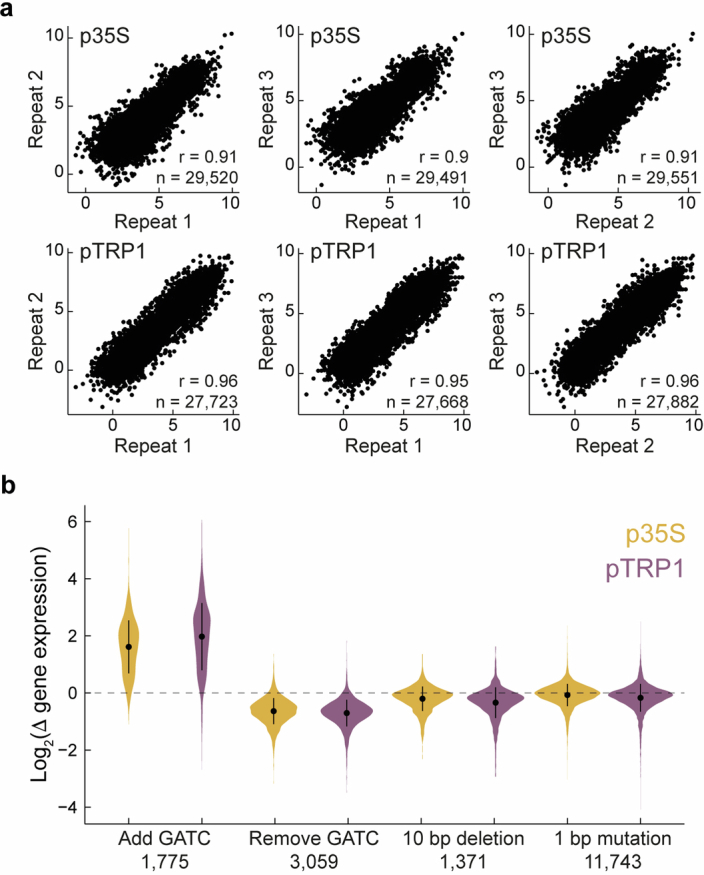
MPRA experiments were performed in Arabidopsis protoplasts using synthetically mutated sequences inserted only in the downstream position. Both p35S- and pTRP1-based libraries were used. The libraries contained 30,000 fragments: 12,000 from the initial pool and an additional 18,000 fragments, each being a variant of one of the original fragments. (a) Comparison between each pair from the three replicates, displaying results with the p35S-based library at the top and the pTRP1-based library at the bottom. Pearson’s correlation coefficients (r) and numbers of compared fragments (n) are indicated on each graph. (b) Log2 expression ratio between mutated fragments and their original fragment. Mutations encompass: addition of a GATC motif, removal of a GATC motif, 10 bp deletions, and 1 bp changes, which include both deletions and nucleotide substitutions. Color represents the library type, either p35S- or pTRP1-based; numbers below the x-axis indicate the number of fragments in each category. Error bars depict the mean with ±1 standard deviation.
