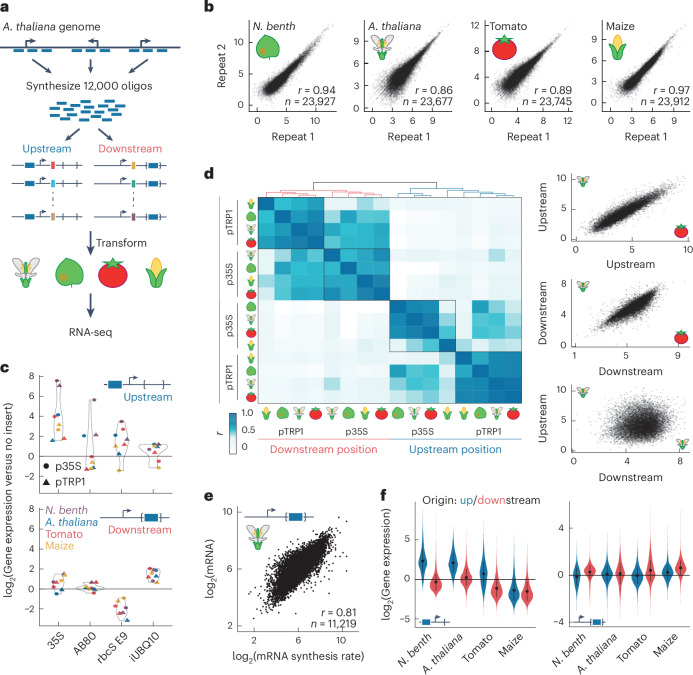
Fig. 2. Position-dependent enhancers reside inside transcribed regions.
a, MPRA overview: 12,000 fragments (160 bp), originating from upstream or downstream of the TSS of Arabidopsis genes, were synthesized, pooled and inserted upstream of the TSS or within the intron of a reporter gene and tagged with barcodes. Following transient transformation into one of four species, barcoded RNA sequencing was used to quantify expression. b, High reproducibility in MPRA experiment replicates, demonstrated here for CaMV 35S minimal promoter-based libraries, plotted as log2(gene expression). Pearson’s correlation coefficient r and number of fragments n are indicated. c, Comparison of construct expression with control enhancer fragments and no-insert constructs for upstream (top) and downstream (bottom) insertions. Depicted for N. benthamiana (N. benth; purple), A. thaliana (blue), tomato (red) and maize (yellow); construct backgrounds are coded by symbol shape. d, Left: Pearson’s correlation coefficients across all libraries, hierarchically clustered. Construct background and insertion position are indicated. Right: activity of pTRP1-based constructs, as log2(gene expression), compared between upstream (top, r = 0.92, n = 11,966) and downstream (middle, r = 0.85, n = 11,817) libraries of Arabidopsis and tomato and between upstream and downstream libraries of Arabidopsis (bottom, r = 0.13, n = 11,813). e, Comparison of mRNA steady-state levels and synthesis rates in the downstream MPRA with pTRP1 constructs. Correlation and fragment counts are indicated as in b. f, Comparison of activity of upstream-derived (blue, 3,966 fragments) and downstream-derived (red, 7,928 fragments) fragments relative to no-insertion constructs when fragments are positioned upstream (left) or downstream (right) of the TSS. Constructs are p35S based. Error bars depict mean ± 1 s.d.