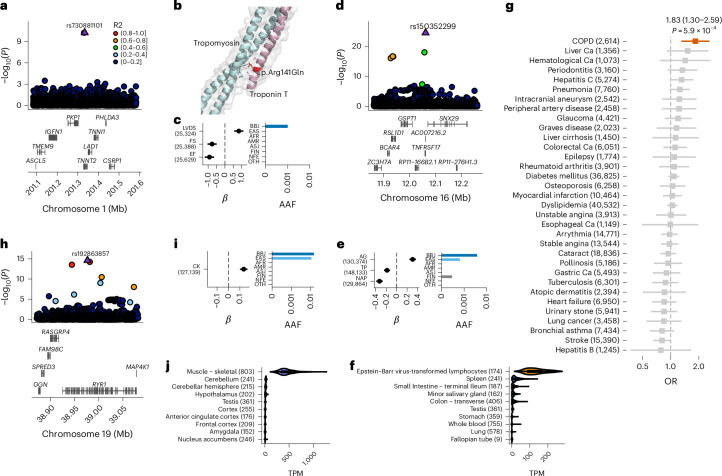
Fig. 1. New rare putative causal coding variants associated with human quantitative traits implicate candidate causal genes.
a, Deleterious coding variant in TNNT2 (rs730881101) showing strong associations with cardiac functions. The horizontal axis indicates the genomic coordinates; the vertical axis indicates the negative log10(P). Statistical significance was tested using a linear mixed model. The displayed P values are two-sided and not adjusted for multiple testing. b, Three-dimensional structure of Troponin-T and putative effect of the coding variant. c, β estimates, PPI and alternative allele frequency (AAF) of rs730881101. The error bar for the β estimates indicates the 95% CI. The number of individuals included in the analysis is shown after the trait names. d, A deleterious coding variant in TNFRSF17 (rs150352299) showing strong associations with AG ratio and non-ALB protein levels. The horizontal axis indicates the genomic coordinates; the vertical axis indicates the negative log10(P). e, β estimates, PPI and AAF of rs150352299. f, Bulk tissue expression of TNFRSF17 in the GTEx. The number of samples is shown after the organ name. The violin plots show the distribution of gene expression in transcripts per million (TPM). The box plot shows the median value as the centerline; the box boundaries show the first and third quartiles and the whiskers extend 1.5 times the interquartile range. g, OR for 29 diseases of rs150352299 in unrelated Biobank Japan (BBJ) participants. Case counts are shown after the outcomes (nTotal = 169,020). The squares indicate the OR; the error bars indicate the 95% CI. Statistical significance was tested using a logistic regression with two-sided test at P < 0.05/29. The displayed P values were not adjusted for multiple testing. h, Deleterious coding variant in RYR1 (rs192863857) associated with CK levels. i, β estimate, PPI and AAF of rs192863857. j, Bulk tissue expression of RYR1 in the GTEx. AFR, African; AMR, Admixed American; ASJ, Ashkenazi Jewish; Ca, cancer; FIN, Finnish; NFE, non-Finnish European; OTH, others. The AAF was obtained from the Genome Aggregation Database (gnomAD) dataset. The number of individuals included in the association analysis is found in Supplementary Table 1; the abbreviations for the phenotypes are found in Supplementary Table 2.