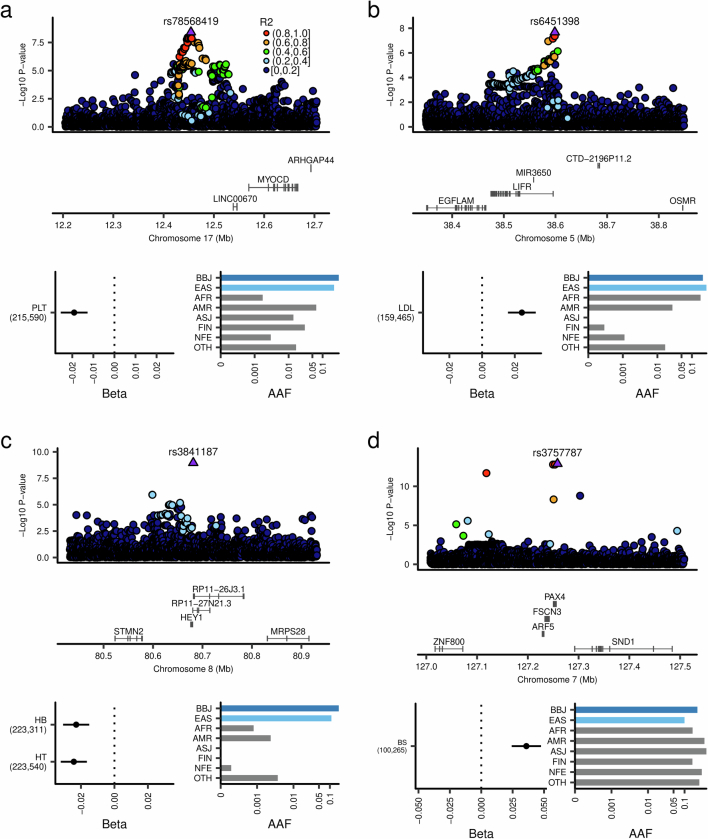
Extended Data Fig. 4. Non-coding variants much more frequent in East Asians than Europeans in novel gene-phenotype pairs.
a, The non-coding variant in the LINC00670 region, rs78568419, which is much more frequent in EAS than EUR, is associated with platelet counts. b, The non-coding variant in the LIFR region, rs6451398, quite rare in Europeans, is associated with LDL levels. c, The non-coding variant in the HEY1 region, rs3841187, which is much more frequent in EAS than the other populations (almost absent in Europeans), showed an association with hemoglobin and hematocrit. d, The non-coding variant in the PAX4 region is associated with blood glucose levels. While this variant is similarly frequent between EAS and EUR, this association was not previously reported. Beta estimates, PPI, and AAF of the associated variants are also indicated in each panel. The error bar for beta estimates indicates 95% confidence interval. The numbers of individuals included in the analysis are shown after the trait names. PPI, posterior probability of inclusion; AAF, alternate allele frequency; BBJ, Biobank Japan; EAS, East Asian; AFR, African; AMR, Admixed American; ASJ, Ashkenazi Jewish; FIN, Finnish; NFE, non-Finnish European; OTH, others. AAF was obtained from the gnomAD dataset. The numbers of individuals included in the association analysis are found in Supplementary Table 1, and abbreviations for phenotypes are found in Supplementary Table 2.