Abstract
The human gut microbiota significantly influences various physiological systems, including immune, nervous, and metabolic systems. Recent studies suggest that gut microbiota may affect sleep quality with certain bacteria and metabolites being linked to sleep patterns. However, the underlying chemical signaling pathway remains unclear. In this study, we investigated the effect of four gut bacteria-derived metabolites, tryptamine (1), indolokine A5 (2), 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE, 3), and phenethylamine (PEA, 4), on sleep characteristics in mice and melatonin biosynthesis pathways in zebrafish. Their sleep-promoting effects were evaluated in a pentobarbital-induced sleep mouse model, revealing significant increases in sleep duration and blood melatonin levels, particularly with ITE (3) and PEA (4). Further tests in zebrafish embryos showed that ITE (3) and PEA (4) increased the expression of genes for melatonin biosynthesis (Aanat1, Aanat2, Tph1a, and Hiomt) in a concentration-dependent manner, indicating their potential as therapeutic agents for sleep disorders.
Introduction
The human gut microbiota has emerged as a crucial factor in maintaining overall health and influencing various diseases. Recent studies have emphasized the significant role that the complex community of microorganisms in the gastrointestinal tract plays various physiological processes, including the immune,1 nervous,2 and metabolic systems.3 This has spurred a growing interest in understanding the intricate interactions between gut microbiota and human health. One area of particular interest is the relationship between gut bacteria and sleep quality. Preliminary research has suggested that the gut microbiota may influence sleep patterns and sleep quality through various mechanisms.4,5 For instance, some butyrate- and lactate-producing bacteria have been linked to sleep characteristics,4,6 and certain gut bacterial metabolites such as benzophenone, pyrogallol, 5-aminopental, and deoxycholic acid have been suggested to be associated with insomnia.7 Despite these insights, the specific chemical signaling pathways through which gut microbiota influence sleep remain largely unexplored.
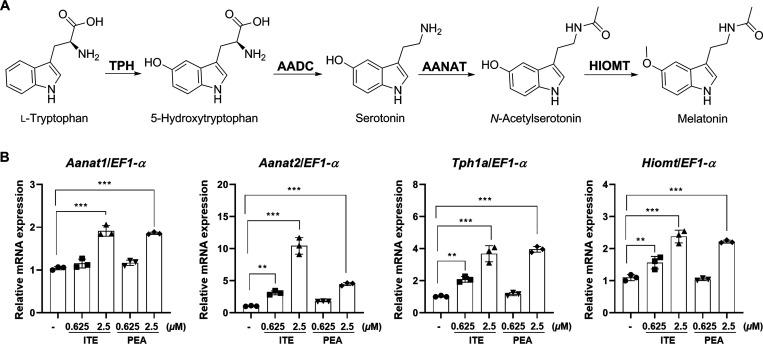
Melatonin, a hormone naturally produced primarily in the pineal gland of the brain, is essential for circadian rhythms, commonly known as the “biological clock”. Melatonin regulates the timing of sleep and wakefulness by influencing over the physiological processes associated with the sleep-wake cycle. Notably, melatonin offers several advantages, including a minimal risk of dependency, absence of habituation, and a generally low incidence of side effects.8 Zebrafish display a remarkable preservation of circadian rhythms governed by melatonin, which is predominantly synthesized in their pineal gland. Given the striking similarity in melatonin synthesis pathways between zebrafish and mammals, a multitude of researchers utilize zebrafish as a valuable model organism for investigating melatonin and sleep patterns.9 The process of melatonin biosynthesis is highly conserved, starting with tryptophan as the precursor. Initially, tryptophan hydroxylase (TPH) catalyzes the conversion of tryptophan to 5-hydroxytryptophan (5-HTP), which is subsequently transformed into serotonin by the enzymatic activity of 5-hydroxytryptophan decarboxylase. Following this, melatonin is synthesized from serotonin through a two-step enzymatic process. In the first step, arylalkylamine N-acetyltransferase (AANAT) facilitates the conversion of serotonin to N-acetylserotonin. AANAT plays a pivotal role in regulating the daily pattern of melatonin production. Notably, zebrafish possess two variants of AANAT, AANAT1 and AANAT2, which are selectively expressed in the retina and the pineal gland, respectively.10 During the nighttime, the activity of AANAT increases significantly by 10- to 100-fold, resulting in an enhanced synthesis and secretion of melatonin. Lastly, hydroxyindole-O-methyltransferase (HIOMT) methylates N-acetylserotonin to form melatonin, thereby regulating sleep-wake patterns and circadian rhythms.11
Enhancing the expression of melatonin biosynthetic enzymes or providing intermediate metabolites involved in the biosynthetic process could potentially lead to improved sleep quality. First, AANAT plays a crucial role in directly controlling the production of melatonin. Consequently, natural or synthetic compounds capable of inducing AANAT expression have the potential to elevate melatonin levels and improve sleep quality. Vitamin A, which primarily functions through its active form, retinoic acid (RA), is a well-known example of such a substance.12 Ashton et al. reported that RA is involved in the induction of AANAT, as part of their investigation into circadian fluctuations in its synthesis and signaling in the rat pineal gland.12 Second, in addition to upregulating the expression of melatonin biosynthetic enzymes, supplying intermediate metabolites is known to enhance sleep quality. 5-HTP is an intermediate substance in the serotonin synthesis pathway derived from tryptophan. It is commonly found in certain dietary supplements and foods and is often used to increase serotonin levels.13 Meloni et al. reported that the consumption of 5-HTP resulted in an increase in REM sleep (rapid eye movement), consequently improving the quality of sleep.14 In addition to 5-HTP, a precursor to 5-HTP, is a commonly used dietary supplement, but it is less effectively absorbed and utilized than 5-HTP. Sutanto et al. reported that the consumption of more than 1 g of l-tryptophan by individuals experiencing insomnia led to an improvement in sleep quality.15 Overall, there is a growing demand for sleep supplements that target the natural melatonin synthesis pathway (AANAT, TPH, and HIOMT), as opposed to prescription sleep medications that can cause serious side effects. Therefore, increasing the genes associated with melatonin biosynthesis to establish the circadian rhythm of the melatonin hormone could serve as a crucial indicator for the development of sleep-regulating agents or novel antidepressant therapies.
Melatonin has an indole functional group in its chemical structure and diverse indole-derived metabolites are produced by commensal gut bacteria including indole, indole-3-pyruvate, indole-3-propionate, indole-3-aldehyde, indole-3-acetate,16 tryptamine,17 and indolokines.18 Among them, indolokines and tryptamine displayed significant agonistic activity on the human aryl hydrocarbon receptor (AhR) at 8 nM (for indolokines).18,19 AhR is a cytoplasmic receptor and a transcription factor in humans that plays important roles in immunity and tissue homeostasis.20 In addition, AhR is highly integrated with the circadian signaling pathways,21−25 however, the underlying molecular mechanisms are largely unknown. Therefore, in this study, we aim to investigate the effects of three gut bacteria-derived AhR agonists tryptamine (1), indolokine A5 (2) and 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE, 3) as well as a structurally related gut bacterial metabolite, phenethylamine (PEA, 4) (Figure 1A), on sleep characteristics in a mouse model and on the genes involved in the melatonin synthesis pathway in a zebrafish model.
Figure 1.
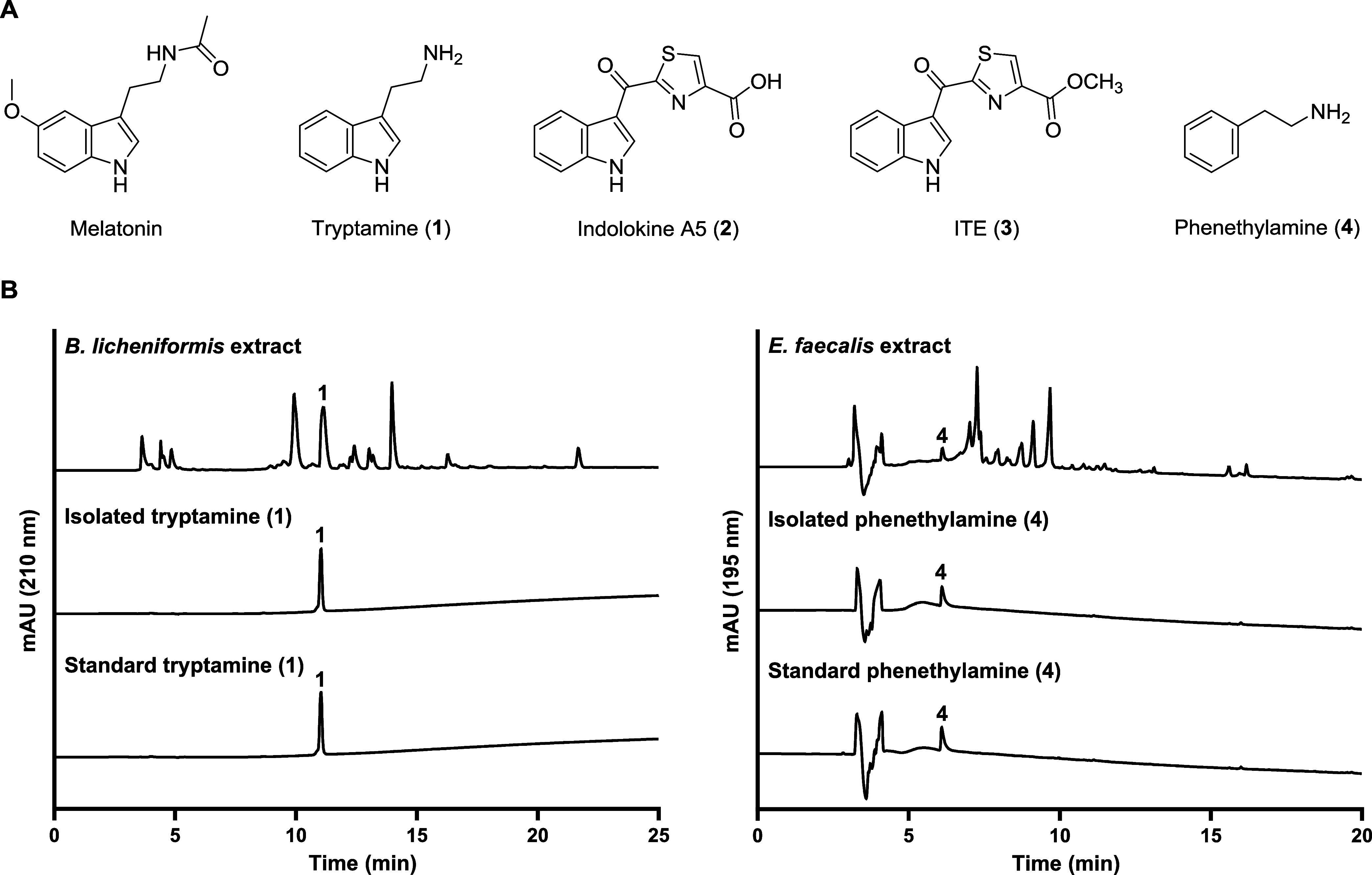
Identification of metabolites from gut bacterial cultures. (A) Structures of melatonin and the gut bacterial metabolites (1–4). (B) HPLC analysis of tryptamine (1) and phenethylamine (4) from bacterial culture extracts (top), isolation (middle), and authentic standards (bottom).
Results and Discussion
Isolation and Chemical Derivatization of Gut Bacterial Metabolites 1–4
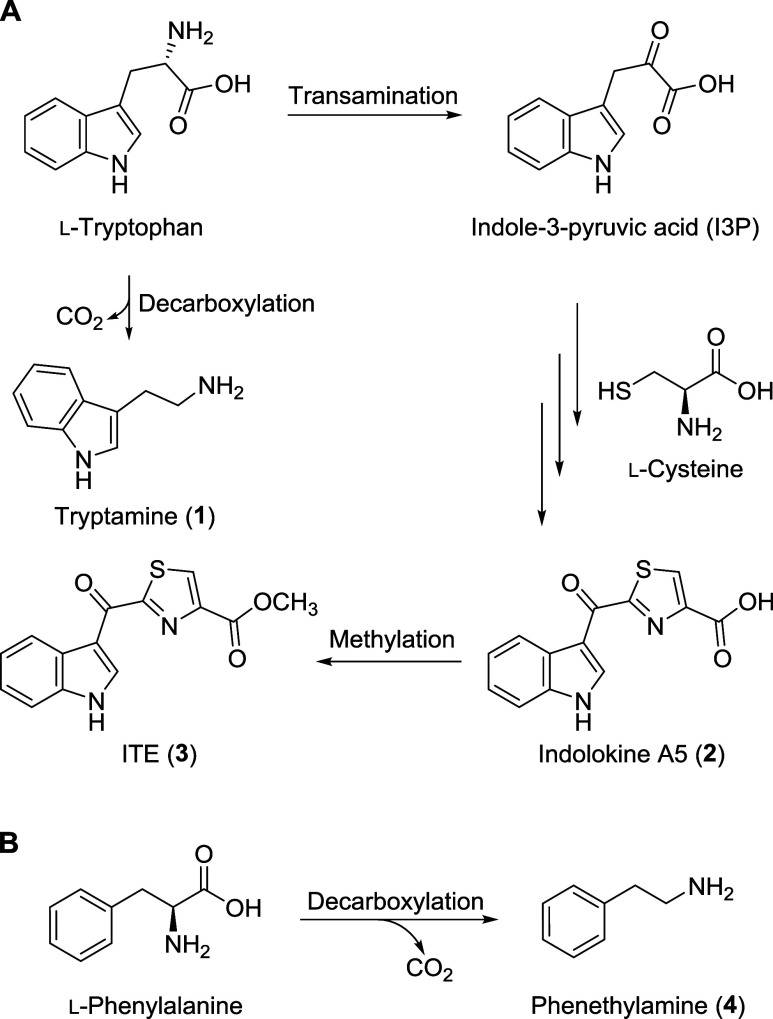
To isolate tryptamine (1) and PEA (4), two gut bacteria, Bacillus licheniformis KCTC 1918 and Enterococcus faecalis KCTC 3206, were cultured in liquid broth, and the metabolites were extracted using EtOAc. The dried organic crude extract was then analyzed using liquid chromatography–mass spectrometry (LC–MS), which identified the two target metabolites 1 and 4 (Figure 1B). The scaled-up cultures of B. licheniformis and E. faecalis were fermented and extracted with EtOAc. Each of the dried extracts was fractionated and purified by HPLC. In the subfraction of the B. licheniformis culture extract, tryptamine (1) was isolated (Yield: 25%, calculated by HPLC). Similarly, PEA (4) was obtained from the culture extract of the E. faecalis (Yield: 3%, calculated by HPLC). The chemical structures of tryptamine (1) and PEA (4) were identified by comparison of their LC-MS data and 1H NMR spectra with previously reported data (Figures S2 and S9).26,27 Tryptamine (1) and PEA (4) are well-known bacterial metabolites, synthesized through the decarboxylation of l-tryptophan and l-phenylalanine, respectively, by various microorganisms (Figure 2).17,28,29 Indolokine A5 (2) was also proposed to be derived from l-tryptophan by transamination and addition of l-cysteine which was confirmed by biomimetic synthesis.18 Through the reported synthetic method, we chemically synthesized indolokine A5 (2) and ITE (3). In addition, we identified indolokine A5 (2) and ITE (3), the methylated analogue of indolokine A5 (2), in E. faecalis culture extracts by LC-MS analysis using EICs with corresponding m/z values and comparing retention times of the natural and synthetic indolokine A5 (2) and ITE (3) (Figures S11 and S12).
Figure 2.
Proposed biosynthesis of gut bacterial metabolites 1–3 (A) and 4 (B).
Sleep-Promoting Effects of Metabolites 1–4
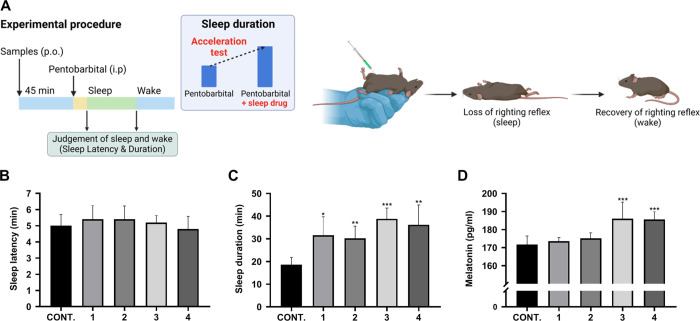
To investigate the sleep-promoting effects of metabolites 1–4, we orally administered each metabolite individually (20 mg/kg) to 10-week-old wild-type male C57BL/6 mice and injected pentobarbital intraperitoneally (42 mg/kg) 45 min later (Figure 3A). We then measured sleep latency until loss of the righting reflex, sleep duration until recovery of the righting reflex, and blood melatonin concentration. As a result, the sleep latency of the control group was 5.0 ± 0.4 min, and no significant change was observed in the metabolite-treated groups (Figure 3B). However, sleep duration was significantly increased in all the tested groups compared to the control group. Metabolites 1–4 showed 31.6 ± 5.6, 30.2 ± 3.5, 38.8 ± 4.8, and 36.2 ± 3.4 min, respectively, which are longer than that of the nontreated control group, 18.6 ± 3.4 min (Figure 3C). We also measured blood melatonin concentration which showed a significant increase of melatonin concentration by ITE (3) and PEA (4) with 186.0 ± 42.9 and 185.6 ± 20.8 pg/mL, respectively, whereas tryptamine (1) and indolokine A5 (2) were not active (Figure 3D). In summary, we found that ITE (3) and PEA (4) are the potent metabolites that contribute to both the extension of sleep duration and increase in blood melatonin concentrations. Therefore, we further tested these two metabolites to understand how they regulate melatonin production at the genetic level.
Figure 3.
Evaluation of the sleep-promoting effects of 1–4 in a pentobarbital-induced sleep mouse model. (A) The experimental procedure for the pentobarbital-induced sleep test method. (B–D) The effect of compounds 1–4 on sleep latency (B), sleep duration (C), and blood melatonin concentration (D) in mice treated with pentobarbital. Error bars represent the mean ± SD. Significance was evaluated using a t-test. Significance levels are indicated as *p < 0.05, **p < 0.01, and ***p < 0.005, compared to the control (CONT.).
Developmental and Heart Toxicity Test of ITE (3) and PEA (4) in Zebrafish Embryos
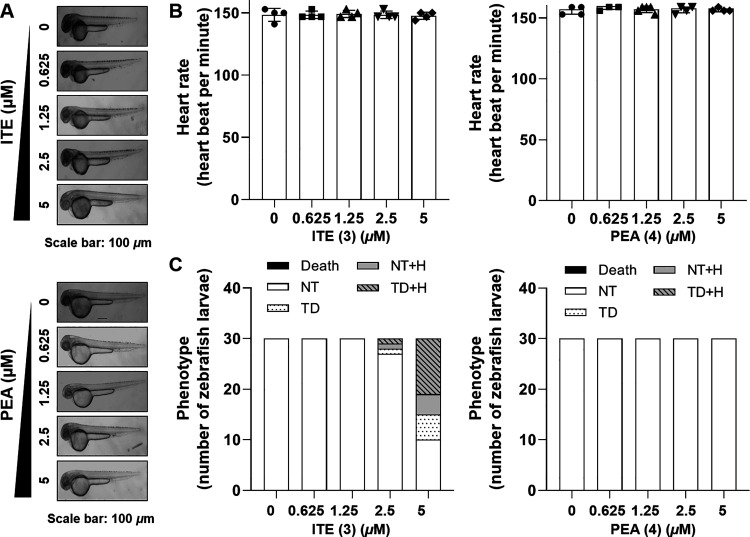
Zebrafish are an ideal model for sleep studies because they exhibit sleep behaviors similar to those in mammals, including characteristic resting postures and reduced sensitivity to stimuli.30 As diurnal vertebrates, they show distinct patterns of daytime activity and nighttime rest, along with melatonin secretion cycles, making them suitable for studying the impact of melatonin on sleep.31 Additionally, zebrafish are valuable for basic toxicity testing due to their genetic similarity to humans, rapid development, and transparent embryos. Their small size, high fecundity, and cost-effectiveness enable efficient toxicological screening, supporting their use in our research. To investigate the toxicity of ITE (3) and PEA (4) in vivo, we observed the average heartbeat rate and phenotypes of zebrafish embryos. When treated with 0.625, 1.25, 2.5, and 5.0 μM ITE (3), no significant effect on the heart rate and developmental progress of zebrafish embryos was observed (Figure 4A–C). PEA (4) also did not show toxicity on heart rate, but some abnormal phenotypes were observed at 5 μM (Figure 4A–C). Based on this, 2.5 μM, a concentration that did not show toxicity, was selected as the high concentration treatment condition for further experiments.
Figure 4.
Assessment of in vivo toxicity of ITE (3) and PEA (4). (A) At the 48 hpf, illustrative photomicrographs were acquired. (B) Zebrafish heartbeat per min. (C) Zebrafish phenotype, classified as normal trunk (NT), trunk deformation (TD), NT with hemorrhage (H), TD with H, and death, was quantified to represent the quantity of zebrafish embryos (n = 30).
ITE (3) and PEA (4) Increase the Melatonin Biosynthesis Pathway in Zebrafish Embryos
Melatonin (5-methoxy-N-acetyltryptamine) is a biogenic hormone secreted by the pineal gland in the brain. It is widely used as a drug to treat sleep disorders or insomnia. Melatonin is synthesized from tryptophan by a four-step enzymatic reaction involving TPH, AANAT, HIOMT (Figure 5A). Being a naturally occurring hormone in the body, strategies to enhance the gene expressions related to melatonin biosynthesis are less likely to have side effects and prove more effective than using artificial sleep aids. To assess the effects of ITE (3) and PEA (4) on sleep, we measured the expression levels of key enzymes in the melatonin biosynthetic pathway, including Tph1a, Aanat1, Aanat2, and Hiomt in zebrafish embryos in vivo. As shown in Figure 5B, treatment with ITE (3) and PEA (4) led to a concentration-dependent increase in the expression of melatonin biosynthetic enzymes, Aanat1, Aanat2, Tph1a, and Hiomt, compared to the control group. The increased expression of these melatonin biosynthetic enzymes is closely linked to the elevation in melatonin production, indicating that ITE (3) and PEA (4) have the potential to be developed as therapeutic agent for the prevention, improvement, and treatment of sleep disorders.
Figure 5.
Evaluation of the effects of ITE (3) and PEA (4) on melatonin biosynthesis. (A) Characterization of the biosynthetic pathway of melatonin. (B) The enhancing effects of ITE (3) and PEA (4) on melatonin biosynthesis in zebrafish embryos. The relative expression levels of Aanat1, Aanat2, Tph1a, and Hiomt mRNA in zebrafish embryos treated with 0.625 or 2.5 μM ITE (3) and PEA (4). Results are presented as means ± SD (n = 3). Statistical significance was determined using one-way ANOVA followed by multiple comparison tests. Significance levels are indicated as **p < 0.05, and ***p < 0.001.
Discussion
Insomnia, a sleep disorder characterized by difficulty falling asleep or staying asleep, has witnessed a global surge in prevalence. Consequently, the demand for sleep aids has surged as well. Sleep aids, approved by the United States Food and Drug Administration (FDA), can be classified into benzodiazepine-based, nonbenzodiazepine-based, and melatonin receptor agonist medications. Benzodiazepine-based sleep aids enhance γ-aminobutyric acid (GABA) inhibition by increasing the permeability of chloride ions across cell membranes during neuronal excitation, rendering them effective in short-term treatment. However, they alter sleep architecture by reducing slow-wave sleep (SWS) and rapid eye movement (REM) sleep, while increasing stage 2 sleep, leading to reduced sleep depth and next-day grogginess. Moreover, adverse effects encompass cognitive impairment, anterograde amnesia, and psychomotor retardation.32 Similarly, nonbenzodiazepine-based sleep aids, although structurally different, target GABA receptors akin to their benzodiazepine counterparts, exhibiting analogous effects. Yet, these too elicit drowsiness, dizziness, and sleep-related behaviors. Given the substantial drawbacks associated with conventional sleep aids, there is a growing need for alternative treatments.32 Melatonin, a hormone secreted by the pineal gland, plays a crucial role in regulating sleep-wake cycles. It acts through melatonin receptors (MT1 and MT2), modulating sleep latency and circadian rhythms. Unlike benzodiazepine-based or nonbenzodiazepine-based sleep aids, melatonin receptor agonists present a more promising profile. They induce drowsiness by reducing sleep onset latency through interaction with MT1 receptor, while the involvement of MT2 receptor aids in circadian rhythm synchronization. Notably, melatonin receptor agonists avoid the grogginess associated with benzodiazepine-based sleep aids and the behavioral aberrations seen with nonbenzodiazepine-based options. Additionally, they do not carry the risk of dependence or withdrawal.33 Considering these advantages, there is a growing impetus for research into agents that modulate the melatonin pathway to enhance sleep regulation. Such agents could potentially address the limitations of existing insomnia treatments while offering a safer and more effective alternative for individuals struggling with sleep disturbances. Further exploration of melatonin pathway modulators is thus warranted to meet the evolving demands of insomnia management.
There are examples of drugs that modulate the synthesis pathway of melatonin. 5-HTP serves as both a pharmaceutical drug and a naturally occurring component in certain dietary supplements. It serves as a precursor to the melatonin synthesis pathway, which is derived from tryptophan through the enzymatic action of TPH. Following decarboxylation, it leads to the production of serotonin (5-HT). 5-HT plays a role in various physiological processes,34 encompassing sleep, thermoregulation, learning and memory, pain perception, social behavior,35 sexual activity, feeding, motor functions, and biological rhythms.36 The hormone melatonin, primarily secreted by the pineal gland and involved in regulating the sleep-wake cycle, is further synthesized from 5-HT. Thus, 5-HTP plays a pivotal and essential role in the production of key molecules such as 5-HT and melatonin.13
The diurnal rhythm of melatonin, with low levels during the day and elevated levels at night, can be a crucial physiological factor in the initiation and maintenance of human sleep. The period of increased melatonin secretion in the pineal gland coincides with human habitual sleep times,37 and there is a strong correlation between the onset of melatonin secretion and the beginning of evening drowsiness.38 We have identified a simple vertebrate model to investigate the influence of melatonin on sleep. Furthermore, as our data have shown that ITE (3) and PEA (4) play a role in enhancing melatonin synthesis pathways, we intend to investigate in the future the extent to which ITE (3) and PEA (4) contribute to quantitative changes in melatonin production.
Recent research has been actively delving into the intricacies of the proteins involved in melatonin synthesis. AANAT has been a subject of interest due to the presence of an Internal Ribosome Entry Site (IRES) activity in the upstream region of its mRNA (mRNA). This IRES activity plays a crucial role in facilitating the translation of the AANAT protein. When a protein called hnRNP Q binds to this region, it triggers activation, resulting in an upsurge in AANAT enzyme levels. The subsequent increase in this enzyme leads to heightened melatonin production, ultimately promoting sleep.39 Consequently, it is envisioned that future endeavors may explore novel drugs targeting the IRES activity or hnRNP Q associated with these AANATs.
In summary, our data suggest that ITE (3) and PEA (4) have shown concentration-dependent increases in the expression of genes associated with melatonin biosynthesis pathways, including Aanat1, Aanat2, Tph1a, and Hiomt. Further research is warranted to explore the impact of these compounds on sleep patterns and to uncover the underlying molecular mechanisms that regulate the expression of enzymes involved in melatonin synthesis.
Experimental Section
General Experimental Procedures
Nuclear magnetic resonance (NMR) spectra (1H) were analyzed using a Bruker AVANCE III HD 700 NMR spectrometer equipped with a 5 mm TCI CryoProbe at 700 MHz (1H), with chemical shifts given in ppm (δ) (Bruker, Karlsruhe, Germany). The LC-MS analysis was performed on an Agilent 1260 series HPLC system with a diode array detector and a 6130 series ESI mass spectrometer equipped with an analytical Kinetex C18 100 Å column (250 mm × 4.6 mm i.d., 5 μm; flow rate: 0.7 mL/min). Semipreparative high-performance liquid chromatography (HPLC) was conducted using an Agilent 1260 pump, which was equipped with a Luna C18 100 Å column (250 mm × 10 mm i.d., 5 μm; flow rate: 4.0 mL/min) and Luna Phenyl-Hexyl column (250 mm × 10 mm i.d., 5 μm; flow rate: 4.0 mL/min).
Cultivation of B. licheniformis and E. faecalis and Metabolite Analysis
B. licheniformis KCTC 1918 was incubated on tryptic soy agar (TSA) plates for 1 day at 37 °C. A single colony was inoculated into 5 mL of tryptic soy broth (TSB) and cultured for 2 days at 37 °C, 250 rpm. The cultures were extracted with 6 mL ethyl acetate, and the organic solvent layer was evaporated. The dried extracts were dissolved in 200 μL of methanol for HPLC analysis. E. faecalis KCTC 3206 was grown on Luria–Bertani (LB) agar plate at 37 °C for 1 day. A single colony from LB agar plate was inoculated into 5 mL of LB broth and incubated for 2 days at 37 °C, 250 rpm. E. faecalis culture was extracted and sampled in the same way as described above. Metabolites screening was carried out using LC-MS equipped with a Phenomenex Luna 5 μm C18 (2) 100 Å (250 mm × 4.6 mm) column with gradient solvent system (0.7 mL/min, 10–100% aqueous acetonitrile, 0.1% formic acid (F.A), 20 min).
Extraction and Isolation
B. licheniformis KCTC 1918 was grown in six tubes containing 5 mL of TSB broth for 1 day at 37 °C, 250 rpm. The cultures were inoculated to 6 L (1 L × 6) cultures of TSB in six 2.8 L Erlenmeyer flasks and fermented at 37 °C, 250 rpm for 2 days. The 6 L culture was extracted with EtOAc with the volume ratio of 1:1 two times, and the organic layer was evaporated to yield 5.8 g of dried extract. The crude extract (5.8 g) was fractionated into 30 fractions by a semipreparative HPLC system (Phenomenex, Luna 10 μm C18 (2) 100 Å (250 mm × 10 mm i.d.)) with a gradient elution from 10% to 100% aqueous MeCN with 0.01% TFA over 30 min (flow rate: 4 mL/min). Compound 1 (tR 10.0 min, 0.7 mg) was isolated from fraction 6 using semipreparative HPLC (Phenomenex, Luna 10 μm Phenyl-Hexyl 100 Å, 250 mm × 10 mm i.d.) with a gradient system of 10–60% aqueous MeCN with 0.01% TFA over 30 min (flow rate 4 mL/min).
Large-scale fermentation (18 L) of E. faecalis KCTC 3206 was also carried out in 3 L Erlenmeyer flasks contained 1 L of LB medium and were incubated at 37 °C, 250 rpm for 2 days. Extraction with EtOAc and fractionation by a semipreparative HPLC were performed in the same protocol as described above, thereby generating 1.9 g of crude extract and 30 fractions. Compound 4 (tR 13.7 min, 0.5 mg) was isolated from fraction 7 using semipreparative HPLC (Phenomenex, Luna 10 μm C18 100 Å, 250 mm × 10 mm i.d.) with a gradient system of 5–6% aqueous MeCN with 0.01% TFA over 45 min (flow rate 4 mL/min).
Detection of Indolokine A5 (2) and ITE (3) in E. faecalis
E. faecalis KCTC 3206 cultures grown for 1 day in LB broth (5 mL) were used to inoculate the 5 mL of LB media supplemented with l-cysteine (1 mM, Sigma-Aldrich), indole-3-pyruvic acid (1 mM, Sigma-Aldrich), or l-tryptophan (1 mM, Sigma-Aldrich) at a 1:200 dilution. These cultures were fermented at 37 °C, 250 rpm for 2 days. The cultures were extracted with EtOAc at room temperature and the organic layer was evaporated. The dried samples were reacted in methanol with 1 N HCl at 100 °C for 1 h. After the organic solvents were evaporated, the dried extracts were resuspended with 1 mL of methanol and centrifuged. 100 μL of supernatant was used for LC-MS analysis. Esterification reaction with acid confirmed the presence of ITE (3) in the samples.
Pentobarbital-Induced Sleep Test
Wild-type male C57BL/6 mice, approximately 10-week-old, were fasted for 24 h prior to the experiment, with 8 animals per group. The experiment was conducted within a certain period of time between 1:00 PM and 6:00 PM. All experimental compounds (1–4) were administered orally 45 min before intraperitoneal injection of pentobarbital (20 mg/kg), and the pentobarbital was injected at a sleep-inducing threshold concentration (42 mg/kg). After treatment, each mouse was placed in a separate chamber, and sleep latency, sleep duration, and blood melatonin concentration were measured. Sleep latency was evaluated as the time until the loss of righting reflex for more than 1 min after injection of pentobarbital. Sleep duration was assessed as the time taken for the recovery of righting reflex. To measure the blood melatonin concentration, blood samples were collected 30 min after pentobarbital injection, and serum was separated. The separated serum was analyzed using the mouse melatonin ELISA kit (LSBio), employing a competitive EIA method.40 We carried out all the animal experiments by animal experiment guideline (Institutional Animal Care and Use Committee, IACUC)
Fish Maintenance and Care
The Institutional Animal Care and Use Committee (IACUC) of Sungkyunkwan University, South Korea, approved all procedures. AB wild-type zebrafish (Danio rerio) were obtained from the Korea Zebrafish Organogenesis Mutant Bank (ZOMB). Approximately 15 zebrafish per 5 L tank were raised at a temperature of 28.5 °C, with a 14:10 h light/dark cycle.
Zebrafish Toxicity Test
Zebrafish embryos at 24 h post fertilization (hpf) were subjected to treatment with ITE (3) and PEA (4) (the targeted compound of interest) at a constant temperature of 28.5 °C. At 48 hpf, the embryos were carefully transferred to a fresh E3 solution, and subsequently, the chorion was gently removed using either a needle or tweezers. The heart rate was then assessed by manually counting the number of heartbeats for 1 min, while the overall phenotype was observed using a microscope (Macrotech, Daejeon, South Korea).
Real-Time Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Zebrafish embryos at 24 h postfertilization (hpf) were treated for 3 days with ITE (3) and PEA (4) at concentrations of 0.625 and 2.5 μM, respectively. Then, total cellular RNA was isolated from zebrafish using TRIzol (Invitrogen, CA) according to the manufacturer’s instructions. Real-time RT-PCR was performed using a Rotor-Gene SYBR Green PCR kit from Qiagen (Valencia, CA) and oligonucleotide primers were purchased from Macrogen (Daejun, Korea). Real-time reverse transcription-polymerase chain reaction (RT-PCR) was conducted using a thermal cycler with the following conditions: an initial denaturation step at 95 °C for 2–3 min to ensure complete denaturation of the DNA template, followed by 40–45 cycles of denaturation at 95 °C for 15 s and a combined annealing/extension step at 60 °C for 30 s. During the annealing/extension phase, the fluorescence data were collected in order to monitor the amplification of the target DNA in real time. The amplification products from the real-time PCR were quantified using the delta–delta Ct method and normalized to the expression level of elongation factor 1-α (EF1-α). The primers utilized are presented in tabular form in the Supporting Information (Table S1).
Data Analysis and Statistics
All data were presented as the mean ± standard deviation. Statistical significance was determined at p < 0.05. Statistical analysis was conducted using GraphPad Prism 6 (GraphPad Software, Inc., CA). Differences between control and treatment groups were assessed using t-tests or one-way ANOVA, followed by Tukey’s posthoc test. Each experiment was performed in triplicate and repeated at least 3 times.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2021R1C1C1011045, 2022R1A6A1A03054419, RS-2024-00350385), by a grant (21153MFDS607) from the Ministry of Food and Drug Safety of South Korea in 2021–2025, and by the BK21 FOUR Project. We thank Minji Kim (Sungkyunkwan University) for her valuable help in editing and proofreading the language of this paper. Figure 3 and TOC graphic were created with BioRender.com.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.4c06923.
Primer list; a detailed analytical data of characterized compounds: UV–vis, MS, 1H NMR data and purity of 1–4; and EIC chromatograms of 2, 3 (Table S1 and Figures S1–S16) (PDF)
Author Contributions
○ J.-H.L. and S.J.H. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Clemente J. C.; Ursell L. K.; Parfrey L. W.; Knight R. The impact of the gut microbiota on human health: an integrative view. Cell 2012, 148 (6), 1258–1270. 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson T. R.; Debelius J. W.; Thron T.; Janssen S.; Shastri G. G.; Ilhan Z. E.; Challis C.; Schretter C. E.; Rocha S.; Gradinaru V.; et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 2016, 167 (6), 1469–1480. 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura V. K.; Faith J. J.; Rey F. E.; Cheng J.; Duncan A. E.; Kau A. L.; Griffin N. W.; Lombard V.; Henrissat B.; Bain J. R.; et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013, 341 (6150), 1241214 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estaki M.; Langsetmo L.; Shardell M.; Mischel A.; Jiang L.; Zhong Y.; Kaufmann C.; Knight R.; Stone K.; Kado D. Association of subjective and objective measures of sleep with gut microbiota composition and diversity in older men: The Osteoporotic Fractures in Men Study. J. Gerontol.: Ser. A 2023, 78 (10), 1925–1932. 10.1093/gerona/glad011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.; Liang T.; Jiang T.; Li Y.; Yang L.; Wu L.; Yang J.; Ding Y.; Wang J.; Chen M.; et al. Gut microbiota: Candidates for a novel strategy for ameliorating sleep disorders. Crit. Rev. Food Sci. Nutr. 2023, 1–17. 10.1080/10408398.2023.2228409. [DOI] [PubMed] [Google Scholar]
- Yu M.; Chen X.; Huang X.; Gao X. Assessing the causal association between sleep apnea and the human gut microbiome composition: A two-sample Mendelian randomization study. SAGE Open Med. 2024, 12, 20503121241248044 10.1177/20503121241248044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even C.; Magzal F.; Shochat T.; Haimov I.; Agmon M.; Tamir S. Microbiota Metabolite Profiles and Dietary Intake in Older Individuals with Insomnia of Short vs. Normal Sleep Duration. Biomolecules 2024, 14 (4), 419 10.3390/biom14040419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliwise D. L.; Ansari F. P. Insomnia associated with valerian and melatonin usage in the 2002 National Health Interview Survey. Sleep 2007, 30 (7), 881–884. 10.1093/sleep/30.7.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda-Martínez P.; Fernández-Martínez J.; Ramírez-Casas Y.; Guerra-Librero A.; Rodríguez-Santana C.; Escames G.; Acuña-Castroviejo D. The zebrafish, an outstanding model for biomedical research in the field of melatonin and human diseases. Int. J. Mol. Sci. 2022, 23 (13), 7438 10.3390/ijms23137438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelbaum L.; Vallone D.; Anzulovich A.; Ziv L.; Tom M.; Foulkes N. S.; Gothilf Y. Zebrafish arylalkylamine-N-acetyltransferase genes–targets for regulation of the circadian clock. J. Mol. Endocrinol. 2006, 36 (2), 337–347. 10.1677/jme.1.01893. [DOI] [PubMed] [Google Scholar]
- Cazaméa-Catalan D.; Magnanou E.; Helland R.; Besseau L.; Boeuf G.; Falcón J.; Jørgensen E. Unique arylalkylamine N-acetyltransferase-2 polymorphism in salmonids and profound variations in thermal stability and catalytic efficiency conferred by two residues. J. Exp. Biol. 2013, 216 (10), 1938–1948. 10.1242/jeb.080960. [DOI] [PubMed] [Google Scholar]
- Ashton A.; Stoney P. N.; Ransom J.; McCaffery P. Rhythmic diurnal synthesis and signaling of retinoic acid in the rat pineal gland and its action to rapidly downregulate ERK phosphorylation. Mol. Neurobiol. 2018, 55, 8219–8235. 10.1007/s12035-018-0964-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei M. E. 5-Hydroxytryptophan (5-HTP): Natural occurrence, analysis, biosynthesis, biotechnology, physiology and toxicology. Int. J. Mol. Sci. 2021, 22 (1), 181 10.3390/ijms22010181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloni M.; Figorilli M.; Carta M.; Tamburrino L.; Cannas A.; Sanna F.; Defazio G.; Puligheddu M. Preliminary finding of a randomized, double-blind, placebo-controlled, crossover study to evaluate the safety and efficacy of 5-hydroxytryptophan on REM sleep behavior disorder in Parkinson’s disease. Sleep Breathing 2022, 26 (3), 1023–1031. 10.1007/s11325-021-02417-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutanto C. N.; Loh W. W.; Kim J. E. The impact of tryptophan supplementation on sleep quality: a systematic review, meta-analysis, and meta-regression. Nutr. Rev. 2022, 80 (2), 306–316. 10.1093/nutrit/nuab027. [DOI] [PubMed] [Google Scholar]
- Tennoune N.; Andriamihaja M.; Blachier F. Production of indole and indole-related compounds by the intestinal microbiota and consequences for the host: the good, the bad, and the ugly. Microorganisms 2022, 10 (5), 930 10.3390/microorganisms10050930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai Y.; Williams B. B.; Battaglioli E. J.; Whitaker W. R.; Till L.; Grover M.; Linden D. R.; Akiba Y.; Kandimalla K. K.; Zachos N. C.; et al. Gut microbiota-produced tryptamine activates an epithelial G-protein-coupled receptor to increase colonic secretion. Cell Host Microbe 2018, 23 (6), 775–785. 10.1016/j.chom.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C. S.; Li J.-H.; Barco B.; Park H. B.; Gatsios A.; Damania A.; Wang R.; Wyche T. P.; Piizzi G.; Clay N. K.; Crawford J. M. Cellular stress upregulates indole signaling metabolites in Escherichia coli. Cell Chem. Biol. 2020, 27 (6), 698–707. 10.1016/j.chembiol.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.; Jin U.-H.; Allred C. D.; Jayaraman A.; Chapkin R. S.; Safe S. Aryl hydrocarbon receptor activity of tryptophan metabolites in young adult mouse colonocytes. Drug Metab. Dispos. 2015, 43 (10), 1536–1543. 10.1124/dmd.115.063677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde R.; McGaha T. L. The aryl hydrocarbon receptor: connecting immunity to the microenvironment. Trends Immunol. 2018, 39 (12), 1005–1020. 10.1016/j.it.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A. Aryl hydrocarbon receptor (AhR) impairs circadian regulation: Impact on the aging process. Ageing Res. Rev. 2023, 87, 101928 10.1016/j.arr.2023.101928. [DOI] [PubMed] [Google Scholar]
- Jaeger C.; Tischkau S. A. Role of aryl hydrocarbon receptor in circadian clock disruption and metabolic dysfunction. Environ. Health Insights 2016, 10, EHI-S38343 10.4137/ehi.s38343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazaal A. Q.; Haque N.; Krager C. R.; Krager S. L.; Chambers C.; Wilber A.; Tischkau S. A. Aryl hydrocarbon receptor affects circadian-regulated lipolysis through an E-Box-dependent mechanism. Mol. Cell. Endocrinol. 2023, 559, 111809 10.1016/j.mce.2022.111809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischkau S. A. Mechanisms of circadian clock interactions with aryl hydrocarbon receptor signalling. Eur. J. Neurosci. 2020, 51 (1), 379–395. 10.1111/ejn.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger C.; Khazaal A. Q.; Xu C.; Sun M.; Krager S. L.; Tischkau S. A. Aryl hydrocarbon receptor deficiency alters circadian and metabolic rhythmicity. J. Biol. Rhythms. 2017, 32 (2), 109–120. 10.1177/0748730417696786. [DOI] [PubMed] [Google Scholar]
- Baron M.; Métay E.; Lemaire M.; Popowycz F. Reduction of aromatic and aliphatic nitro groups to anilines and amines with hypophosphites associated with Pd/C. Green Chem. 2013, 15 (4), 1006–1015. 10.1039/c3gc37024k. [DOI] [Google Scholar]
- Vatèle J.-M. Prenyl carbamates: preparation and deprotection. Tetrahedron 2004, 60 (19), 4251–4260. 10.1016/j.tet.2004.03.028. [DOI] [Google Scholar]
- Yuwen L.; Zhang F.-L.; Chen Q.-H.; Lin S.-J.; Zhao Y.-L.; Li Z.-Y. The role of aromatic L-amino acid decarboxylase in bacillamide C biosynthesis by Bacillus atrophaeus C89. Sci. Rep. 2013, 3 (1), 1753 10.1038/srep01753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A.; De Las Rivas B.; Landete J. M.; Tabera L.; Muñoz R. Tyramine and phenylethylamine biosynthesis by food bacteria. Crit. Rev. Food Sci. Nutr. 2012, 52 (5), 448–467. 10.1080/10408398.2010.500545. [DOI] [PubMed] [Google Scholar]
- Zhdanova I. V.; Wang S. Y.; Leclair O. U.; Danilova N. P. Melatonin promotes sleep-like state in zebrafish. Brain Res. 2001, 903 (1–2), 263–268. 10.1016/S0006-8993(01)02444-1. [DOI] [PubMed] [Google Scholar]
- Kazimi N.; Cahill G. M. Development of a circadian melatonin rhythm in embryonic zebrafish. Dev. Brain Res. 1999, 117 (1), 47–52. 10.1016/S0165-3806(99)00096-6. [DOI] [PubMed] [Google Scholar]
- Morgenthaler T. I.; Silber M. H. Amnestic sleep-related eating disorder associated with zolpidem. Sleep Med. 2002, 3 (4), 323–327. 10.1016/S1389-9457(02)00007-2. [DOI] [PubMed] [Google Scholar]
- Hardeland R. New approaches in the management of insomnia: weighing the advantages of prolonged-release melatonin and synthetic melatoninergic agonists. Neuropsychiatr. Dis. Treat. 2009, 341–354. 10.2147/NDT.S4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad-Zadeh L. F.; Moses L.; Gwaltney-Brant S. Serotonin: a review. J. Vet. Pharmacol. Ther. 2008, 31 (3), 187–199. 10.1111/j.1365-2885.2008.00944.x. [DOI] [PubMed] [Google Scholar]
- Kravitz E. A. Hormonal control of behavior: amines and the biasing of behavioral output in lobsters. Science 1988, 241 (4874), 1775–1781. 10.1126/science.2902685. [DOI] [PubMed] [Google Scholar]
- Zifa E.; Fillion G. 5-Hydroxytryptamine receptors. Pharmacol. Rev. 1992, 44 (3), 401–458. [PubMed] [Google Scholar]
- Lynch H. J.; Wurtman R.; Moskowitz M. A.; Archer M.; Ho M. Daily rhythm in human urinary melatonin. Science 1975, 187 (4172), 169–171. 10.1126/science.1167425. [DOI] [PubMed] [Google Scholar]
- Åkerstedt T.; Fröberg J. E.; Friberg Y.; Wetterberg L. Melatonin excretion, body temperature and subjective arousal during 64 h of sleep deprivation. Psychoneuroendocrinology 1979, 4 (3), 219–225. 10.1016/0306-4530(79)90005-2. [DOI] [PubMed] [Google Scholar]
- Kim T.-D.; Woo K.-C.; Cho S.; Ha D.-C.; Jang S. K.; Kim K.-T. Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 2007, 21 (7), 797–810. 10.1101/gad.1519507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy J. F.; Dijk D.-J.; Hall E. F.; Czeisler C. A. Relationship of endogenous circadian melatonin and temperature rhythms to self-reported preference for morning or evening activity in young and older people. J. Invest. Med. 1999, 47 (3), 141–150. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.