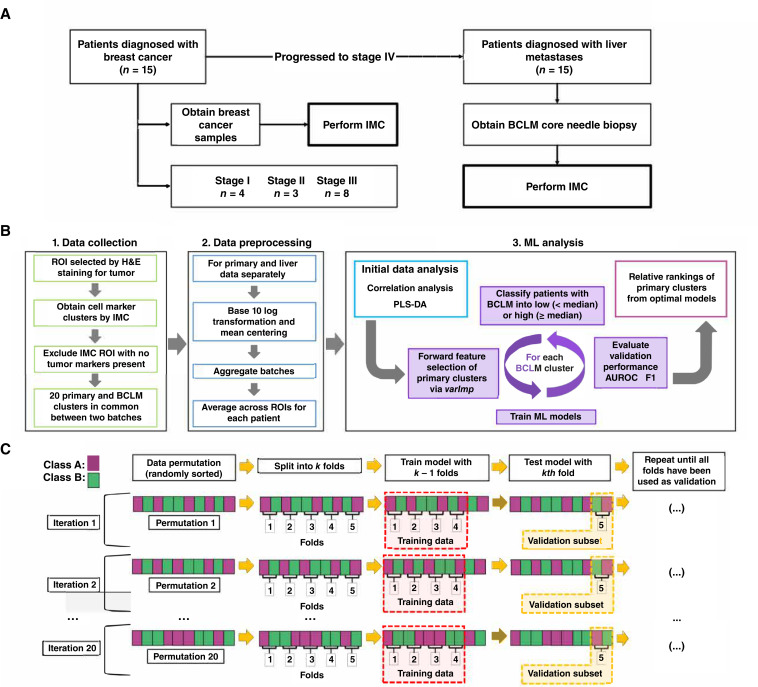
Figure 1.
Workflow of study design. A, Study profile. Primary breast cancer samples were taken from 15 patients. After subsequent diagnosis of BCLM, a core needle biopsy was also obtained. B, Summary of analysis. ROIs from primary tumor and BCLM were identified using H&E staining for tumor tissue and TME. Multiple marker clusters were quantified by IMC, producing 20 common clusters between two analytical batches. IMC ROI missing multiple tumor markers (Ki-67, E-cad+, or αSMA) were excluded. Multiple ML models were trained to classify BCLM cluster expression into low (< median) or high (≥ median) groups using primary tumor cluster data. Forward feature selection was performed on preprocessed data using varImp to identify primary TME markers associated with BCLM classification. C, Diagram of model training and validation. Primary tumor data were randomly sorted and split into k folds (subsets; here, k = 5). Each model was trained with k-1 folds and validated with the kth fold. This process was repeated until all folds were used once as the validation set. Twenty permutations were performed in total, repeating the validation process for each fold within each permutation. Final results of each model are the averages of the validations across all folds and all iterations (n = 100). H&E, hematoxylin and eosin.