Figure 2.
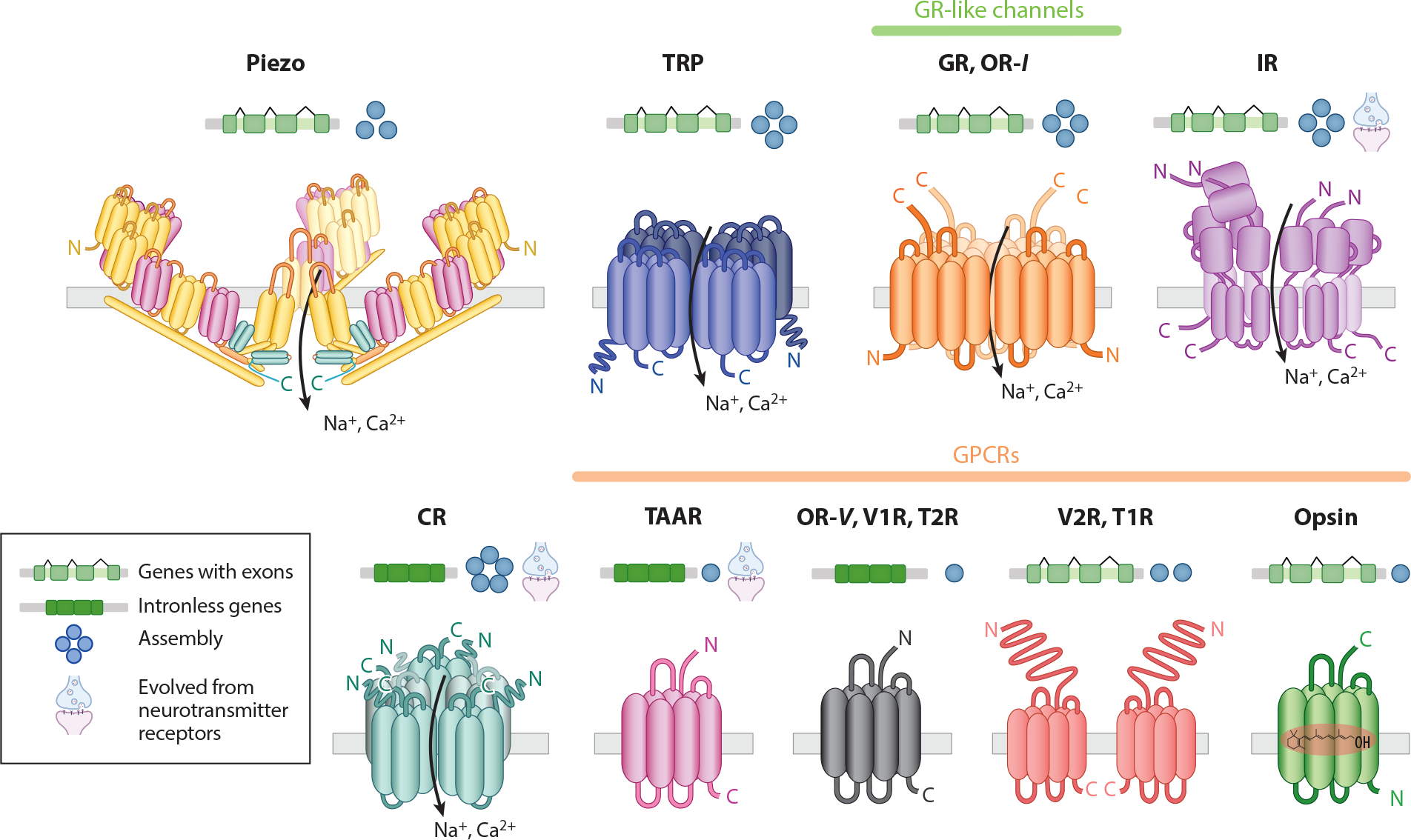
Genomic architecture, protein structure, and membrane topology of sensory receptors, showing patterns of assembly. Exon-intron structures (green) of sensory receptor families hint at the duplication mechanism underlying their evolution. The exon-intron architectures and assemblies were inferred from the majority of the known cases within the gene families. Distinct families across vertebrates and invertebrates diverged from ancestral neurotransmitter receptors, including IRs, CRs, and TAARs. Piezo channels, TRP channels, GRs, ORs-I, IRs, and CRs are nonselective cation channels. GPCRs transduce information through multicomponent second messenger–based signaling pathways. Abbreviations: CRs, chemotactile receptors; GPCRs, G protein–coupled receptors; GRs, gustatory receptors; IRs, ionotropic receptors; ORs-I, insect olfactory receptors; OR-V, vertebrate odorant receptor; T1R, taste receptor type 1; T2R, taste receptor type 2; TAARs, trace amine-associated receptors; TRP, transient receptor potential; V1R, vomeronasal receptor type 1; V2R, vomeronasal receptor type 2.
