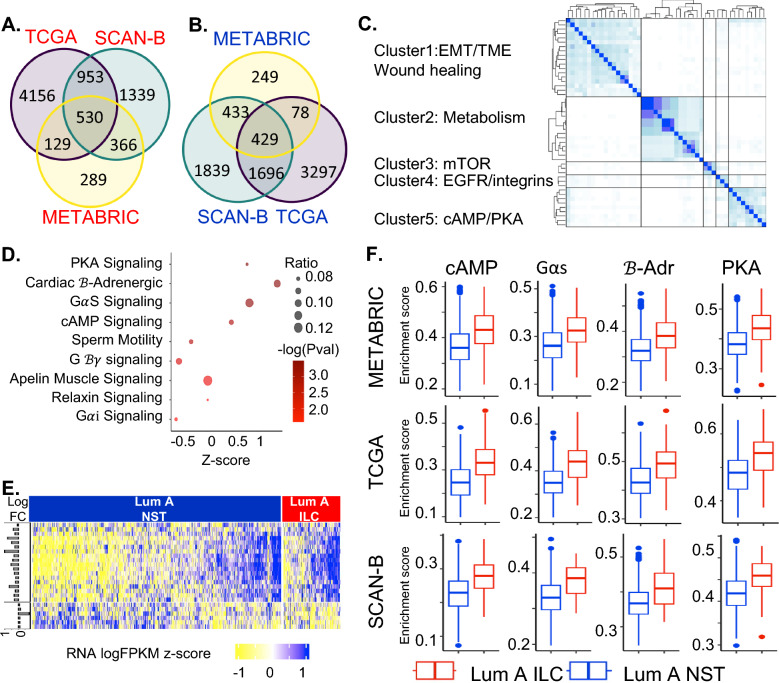
Fig. 2.
A Venn diagram showing overlapping differentially expressed genes upregulated in luminal A ILC (as compared to luminal A NST) in TCGA, SCAN-B, METABRIC. B Venn diagram showing overlapping differentially expressed genes downregulated in luminal A ILC (as compared to luminal A NST) in TCGA, SCAN-B, METABRIC. C Clustering heatmap using Jaccard index for IPA pathways significantly enriched in differentially expressed genes that overlap in all 3 databases; pathways with Jaccard index > 0.3 and at least 1 membership were clustered using hierarchical clustering into clusters of related pathways. D Bubble plot depicting z score (represents overall effect on pathway in Luminal A ILC, z-score > 0 suggests upregulation and z-score < 0 suggests downregulation) in x-axis, -log(P-val) as color and ratio (ratio of differentially expressed genes in the pathway to total number of genes in the pathway) as bubble size, for pathways in Cluster 5 from the Jaccard index heatmap. The reported z-scores, p-values and ratio are based on gene expression data from SCAN-B. E Heatmap showing expression (scaled logFPKM values from SCAN-B) of genes present in the pathways in Cluster 5 of Jaccard index heatmap with a positive z-score (Protein Kinase A Signaling, Cardiac β-Adrenergic Signaling, GαS Signaling, cAMP-mediated Signaling) that are also differentially expressed between Luminal A ILC and Luminal A NST. The horizontal bars on the left depict the log fold change for the corresponding gene in luminal A ILC as compared to luminal A NST in SCAN-B. F Boxplots showing GSVA enrichment score for pathways from Cluster 5 of Jaccard index heatmap with positive z-score (cAMP = cAMP-mediated Signaling, GαS = GαS Signaling, β-Adr = Cardiac β-Adrenergic Signaling, PKA = Protein Kinase A Signaling), in Luminal A ILC (red) and Luminal A NST (blue) in SCAN-B, TCGA and METABRIC; p < 0.0001 for all comparisons. ILC—Invasive lobular breast cancer; NST—Invasive carcinoma—no special type; Lum A—Luminal A; SCAN-B—Sweden Cancerome Analysis Network—Breast; TCGA—The Cancer Genome Atlas; METABRIC—Molecular Taxonomy of Breast Cancer International Consortium; EMT—Epithelial-Mesenchymal Transition; TME—Tumor Microenvironment; mTOR—mammalian Target of Rapamycin; EGFR—Epidermal Growth Factor Receptor; IPA—Ingenuity Pathway Analysis; FPKM—Fragments Per Kilobase of transcript per Million mapped reads; PKA—Protein kinase A; cAMP—cyclic adenosine monophosohate