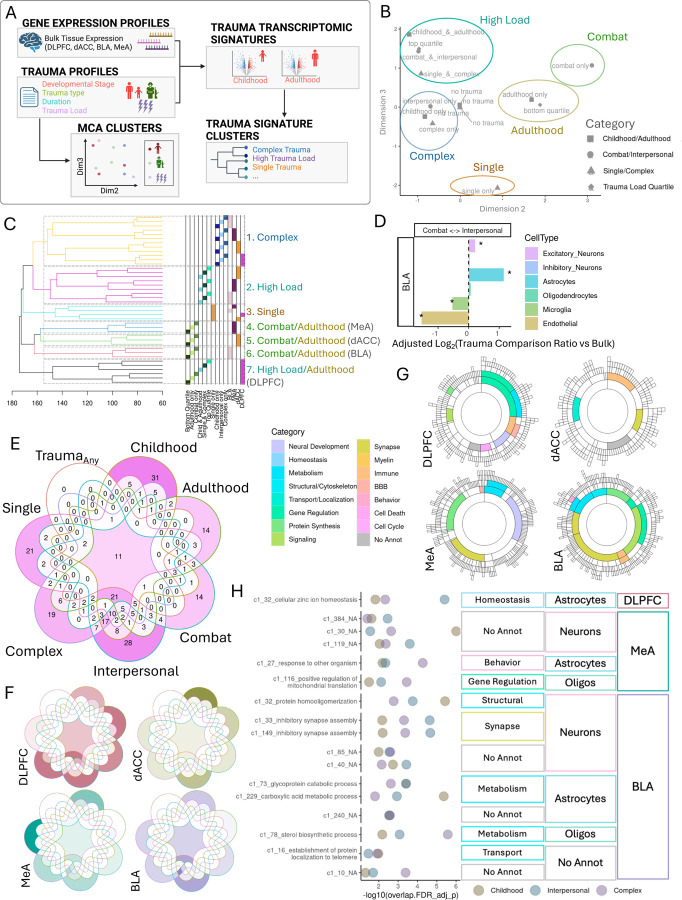
Figure 4.
Trauma types have shared and distinct transcriptional signatures A. Analysis of trauma variable profiles and their transcriptomic signatures. B. Multiple correspondence analysis (MCA) of donor trauma profiles. Dimensions 2 and 3 reveal clusters of similar trauma variable categories, which we categorize into five MCA clusters: High load, complex traumas, adulthood, single and combat. C. Hierarchical clustering of trauma variable category transcriptional signatures. Clustering heights are shown on the x axis. Trauma variable and brain region for each transcriptomic signature is annotated to the right by color and in text with congruent MCA clustering categories. Dashed rectangles designate trauma transcriptional signature clusters. D. Log-transformed ratio of number of interpersonal ctDEGs over number of combat ctDEGs for each cell type (color bar) and adjusted for proportion of bulk interpersonal DEGs over bulk combat DEGs. Significant enrichment of number of interpersonal (positive value) or combat DEGS (negative value) by exact binomial test for deviation from bulk proportion with qFDR< 0.05 (*). E-F. Intersections of the number of modules enriched for each trauma type signature for all brain regions combined (E) and separated by brain region (F). G. Modules commonly enriched for all 6 trauma types (childhood, adulthood, combat, interpersonal, complex and single trauma) and colored by annotated functional category. H. Modules commonly enriched for childhood, interpersonal and complex trauma signatures. Module enrichment log-transformed FDR-adjusted p-value shown for each trauma signature (color). Brain region, cell type and functional annotations shown. Modules are labeled with significant (qFDR< 0.05) GO term with the lowest pvalue.