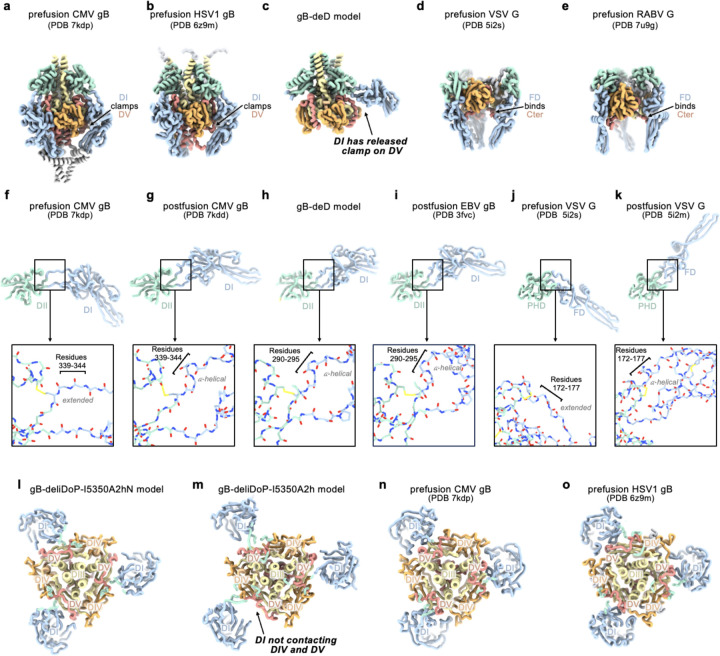
Extended Data Figure 5. Comparing prefusion and postfusion hinge conformations with gB-deD.
Domain I (DI, blue), domain II (DII, green), domain III (DIII, yellow), domain IV (DIV, orange), and domain V (DV, red) are shown as ribbons. (a-e) Side-view comparisons of prefusion CMV gB (a), prefusion HSV1 gB (b), the EBV gB-deD model determined herein (c), prefusion VSV G (d), and prefusion RABV G (e) highlighting how prefusion structures clamp or bind DV (Cter in VSV and RABV G). (f-k) Comparisons of DI hinge structure showing DI and DII (or FD and PHD for VSV) from prefusion CMV gB (f), prefusion HSV1 gB (g), the gB-deD model determined herein (h), postfusion EBV gB (i), prefusion VSV G (j), and postfusion VSV G (k). Insets show zoomed-in view of backbone atoms, plus disulfide bonds and prolines, highlighting the restructuring of prefusion and postfusion DI hinge residues. (l-o) Comparisons of gB cross-sections showing DI in relation to DIV and DV, including EBV gB-deliDoP-I5350A2hN (l), EBV gB-deliDoP-I5350A2h (m), prefusion CMV gB (n), and prefusion HSV1 gB (o).